Machine Learning Services
Machine learning is a branch of artificial intelligence (AI) based on the idea that systems can learn from data, identify patterns and make decisions with minimal human intervention. The major contribution of AI in bioinformatics analyses depends on pattern matching and knowledge based learning systems to solve the biological problems.
Machine learning in Bioinformatics
Before machine learning emerged, bioinformatics and other biological fields faced the problem of extracting valuable insights from large biological datasets. But as of today, ML techniques such as deep learning can learn the features of complex datasets and present them in a manner that is easy to understand.
Machine learning has multiple applications in diverse fields, ranging from natural language processing to healthcare. Machine learning in bioinformatics is the application of machine learning algorithms in bioinformatics; genomics, proteomics, microarrays, systems biology, evolution, and text mining. It serves as an advanced tool in the bioinformatics area which deals with molecular phenotypes, drug discovery, and aids in determining unfamiliar diseases etc.
Molecular Docking Services
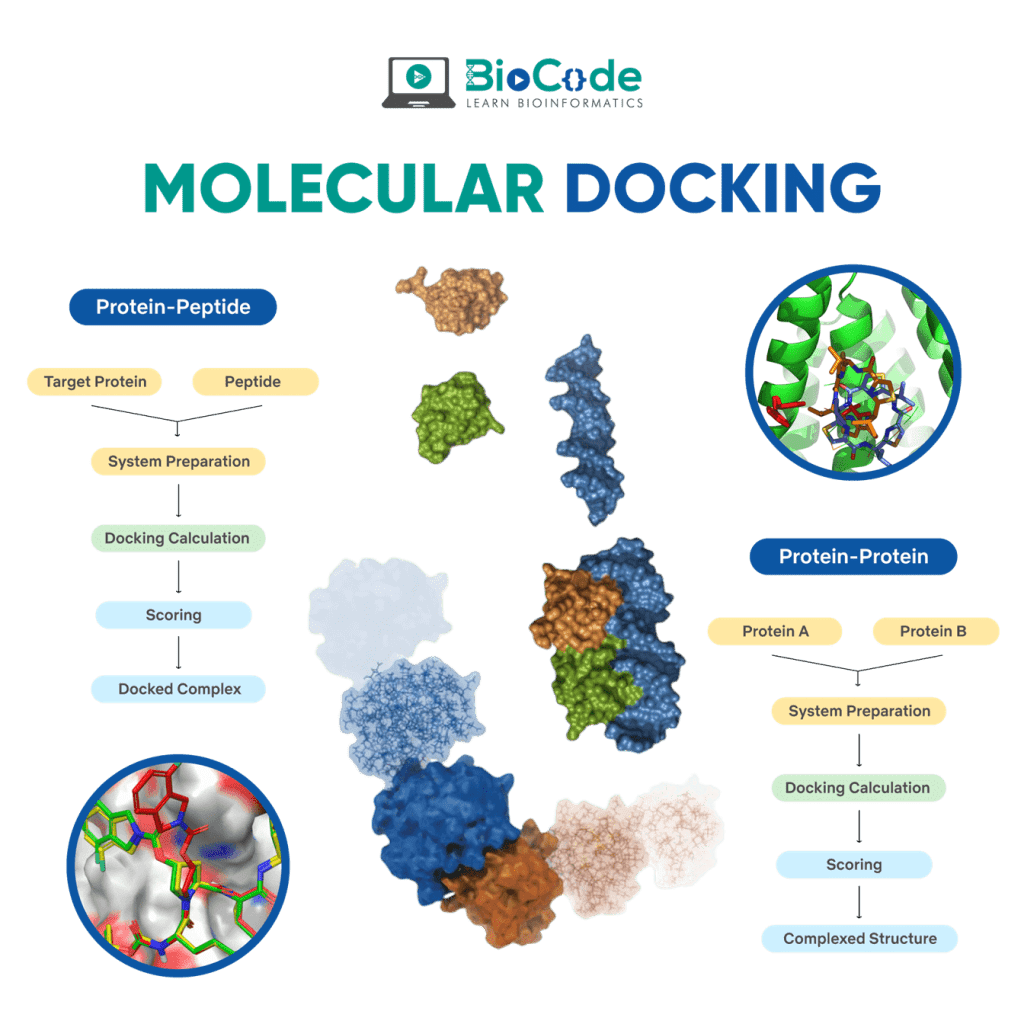
Molecular Docking is a bioinformatics modeling technique which involves the interaction of two or more molecules to give the stable adduct. Molecular docking is an important approach for designing new drugs and vaccines and other bioinformatics analysis as well. It predicts the three-dimensional structure of any complex depending upon binding properties of ligand and target. Molecular docking generates different possible adduct structures that are ranked and grouped together using a scoring function in the software.
Molecular interactions including protein-protein, enzyme-substrate, protein-nucleic acid, drug-protein, and drug-nucleic acid play important roles in many essential biological processes, such as signal transduction, transport, cell regulation, gene expression control, enzyme inhibition, antibody–antigen recognition and even the assembly of multi-domain proteins. These interactions very often lead to the formation of stable protein–protein or protein-ligand complexes that are essential to perform their biological functions.
Molecular Docking Classification
Molecular docking classifies biomolecules into three categories: Proteins, Ligands and Peptides. The most important types of docking include protein-protein docking, protein-ligand docking and protein-peptide docking.
RNA-Seq Analysis Services
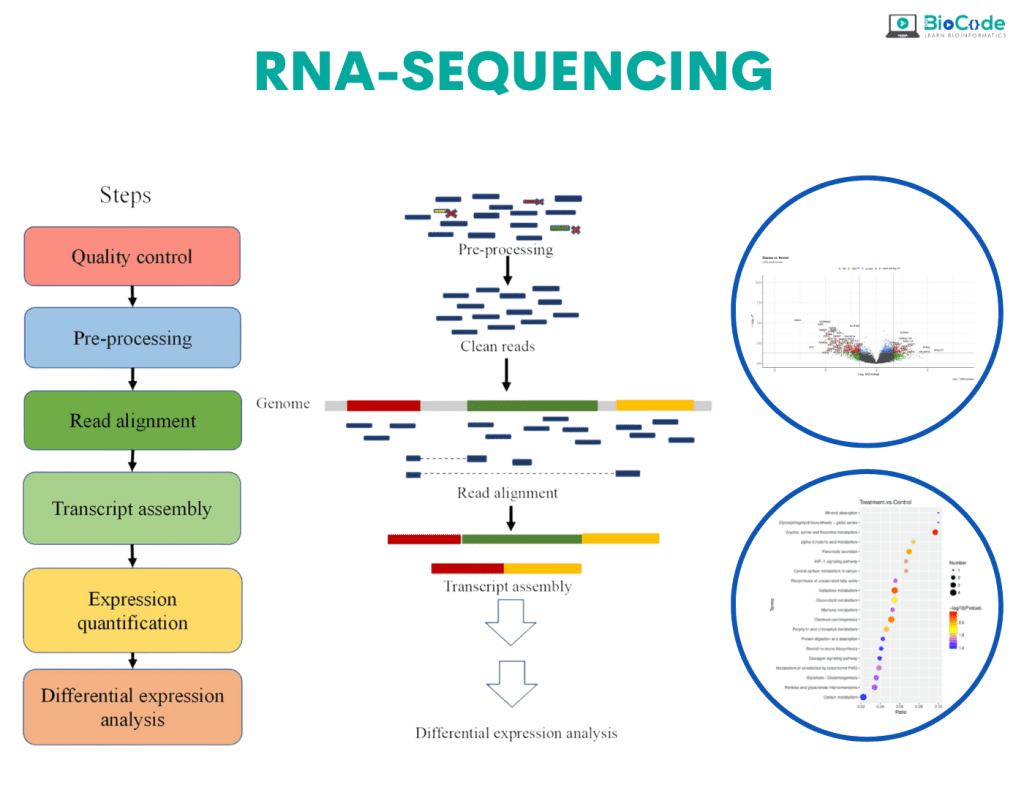
RNA-Seq is an exciting and in-demand next-generation sequencing (NGS) method used for identifying genes and pathways underlying certain diseases or conditions. Over the past decade, RNA-seq has become an indispensable tool for transcriptome-wide analysis of differential gene expression and differential splicing of mRNAs. RNA-seq offers many advantages over microarray technology.
RNA-Sequencing Data Analysis
RNA sequencing data analysis emphasizes the complicated mechanisms of gene regulation. It analyzes the transcriptome, indicating which of the genes encoded in our DNA are turned on or off and to what extent. RNA-seq data allows for a wide range of analyses to address countless research questions across the fields of biology and biomedicine.
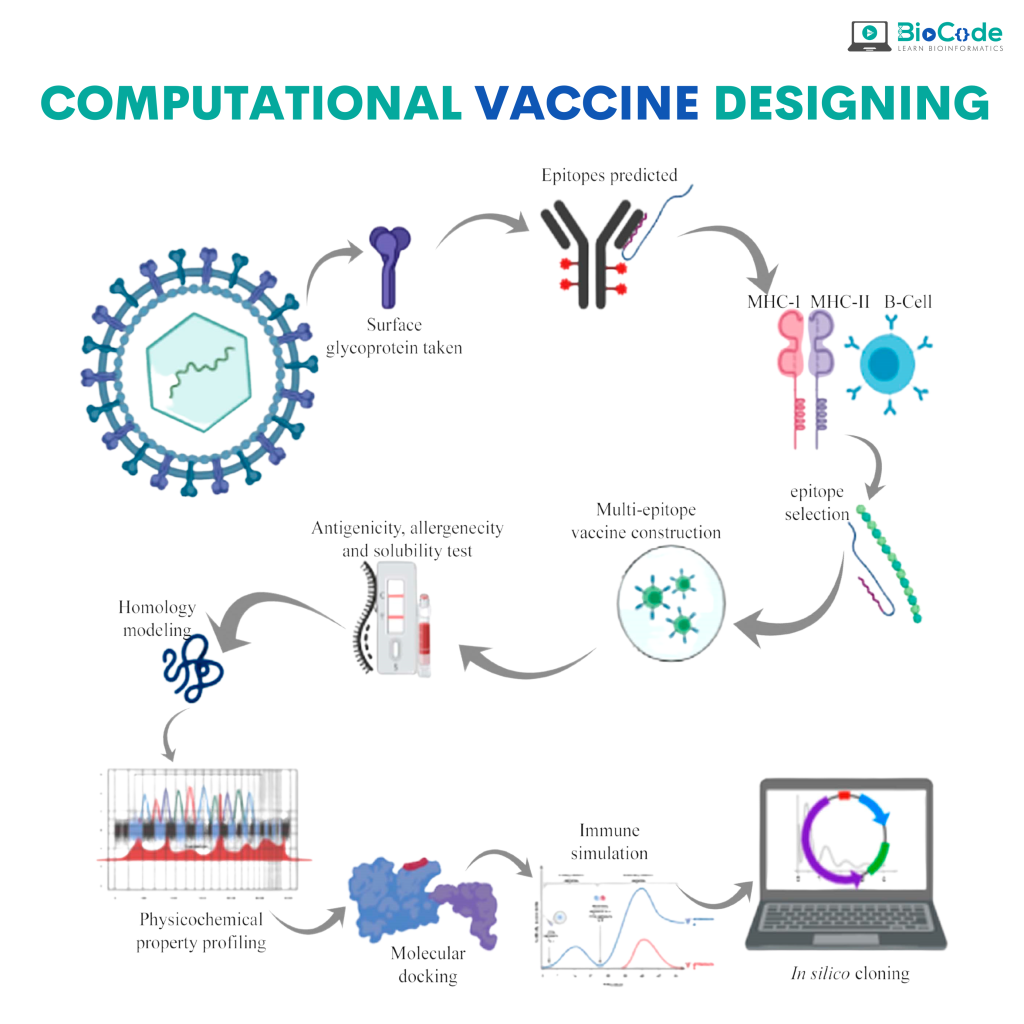
Computational Vaccine Designing and Immunoinformatics Services
Newly emerging and reemerging infectious viral diseases have threatened humanity throughout history. The unprecedented scale and rapidity of dissemination of recent emerging infectious diseases pose new challenges for vaccine developers, regulators, health authorities and political constituencies.
Vaccines are biological substances that are utilized to stimulate antibody production within the body of an organism to provide active immunity against foreign organisms, mostly viruses and bacteria. Vaccines not only arrest the beginning of different diseases but also assign a gateway for its elimination and reduce toxicity. Vaccines are the most cost-effective public health interventions.
Computational Vaccine Designing and Chimeric Vaccines
Bioinformatics is also involved in medication development aimed at bio-productive and pharmaceutical/vaccine development. Computational approach in drug discovery helps in identifying safe and novel vaccines. In silico(computational) analysis saves time, cost, and labor for developing the vaccine and drugs. Chimeric vaccines are types of recombinant vaccines, produced by substituting genes from the target pathogen in a closely related organism, for similar genes. Chimeric vaccines are useful in studying infectious diseases, including many neglected diseases.
Computational Drug Discovery Services
Computational Drug Designing has become the go-to requirement for the researchers, scientists and the pharmaceuticals who fight against the fatal disease. Computational drug discovery is an effective strategy for accelerating and economizing drug discovery and development processes. Because of the dramatic increase in the availability of biological macromolecule and small molecule information, the applicability of computational drug discovery has been extended and broadly applied to nearly every stage in the drug discovery and development workflow, including target identification and validation, lead discovery and optimization and preclinical tests.
Role of Bioinformatics in Drug Discovery
Bioinformatics analysis can not only accelerate drug target identification and drug candidate screening and refinement, but also facilitate characterization of side effects and predict drug resistance. High-throughput data such as genomic, epigenetic, genome architecture, cistromic, transcriptomic, proteomic, and ribosome profiling data have all made significant contributions to mechanism-based drug discovery and drug repurposing. Accumulation of protein and RNA structures, as well as development of homology modeling and protein structure simulation, coupled with large structure databases of small molecules and metabolites, paved the way for more realistic protein-ligand docking experiments and more informative virtual screening.
Molecular Dynamics Simulation Services
Molecular dynamics (MD) is a computer simulation method for analyzing the physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamic “evolution” of the system. In the most common version, the trajectories of atoms and molecules are determined by numerically solving Newton’s equations of motion for a system of interacting particles, where forces between the particles and their potential energies are often calculated using interatomic potentials or molecular mechanics force fields. MD simulations are nowadays routinely applied to macromolecular systems of biological and pharmaceutical interest. MD simulation analysis is one of the essential steps while designing novel drugs using computational approaches.
Molecular Dynamics Simulations in Drug Designing
Atomistic computer simulations of macromolecular (for example, protein) receptors and their associated small-molecule ligands play an essential role in drug discovery. The static models produced by NMR, X-ray crystallography, and 3D structure prediction provide valuable insights into macromolecular structure, but molecular recognition and drug binding are very dynamic processes. When a small molecule like a drug (for example, a ligand) approaches its target (for example, a receptor) in solution, it encounters not a single, frozen structure, but rather a macromolecule in constant motion.
ChIP-Seq Analysis
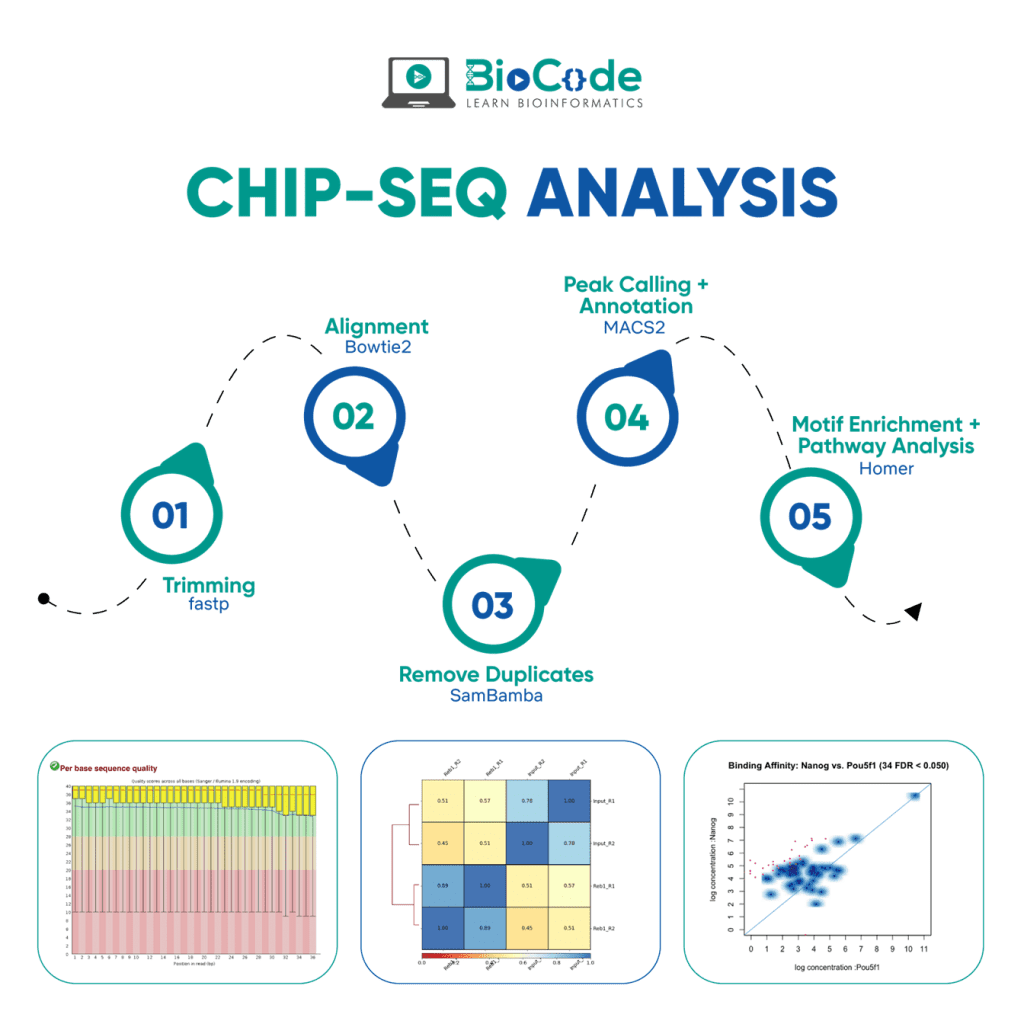
ChIP is an antibody-based technique that is used to enrich specific DNA-binding proteins in addition to their DNA targets. This is used to investigate a specific protein-DNA interaction or multiple protein-DNA interactions across a subset of genes or the whole genome. Chromatin immunoprecipitation (ChIP) assay when combined with sequencing resulted in a powerful high-throughput technique known as the ChIP-Seq.
Chromatin Immunoprecipitation Sequencing (ChIP-Seq)
ChIP-seq is a pivotal technology for epigenomic research and study. ChIP is the method of choice for studying epigenomic signatures. ChIP-seq is different from all of the other approaches, which are used for epigenetic research, in the fact that it does not need any prior knowledge as it does not require probes from known sequences.
ChIP-seq is a powerful method to identify genome-wide DNA binding sites for a protein of interest. ChIP-Seq is an exciting and in-demand next-generation sequencing (NGS) method used for identifying genes and pathways underlying particular diseases or conditions. Through ChIP-Seq Data Analysis you can find protein-DNA interactions, transcription factor binding sites, histone modifications among other epigenetic signatures.
