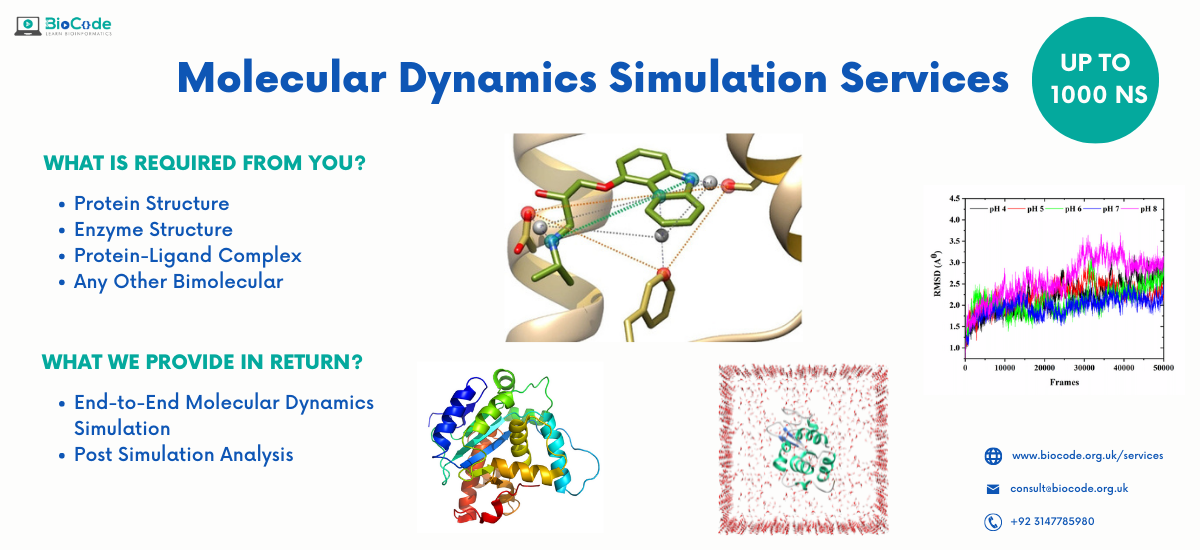
We provide complete services for Molecular Dynamics Simulation Services
From data collection, preprocessing, quality control to transcription factor binding sites, histone modifications and much more.
Providing services to
worldwide clients
Datasets
analyzed
Qualified researchers and
Bioinformatics analysts
Satisfied orders processed
Our Services
- Protein Molecular Dynamics Simulation Service
- Protein-Ligand Docked Complex Simulation Services
- Enzyme Molecular Dynamics Simulation Service
- Lipid Molecular Dynamics Simulation Service
- Membrane Protein Molecular Dynamics Simulation Service
- Calculation of Free Energy (Binding, Solvation, Interaction)
- Interactive Molecular Dynamics
- Visualization Program for Displaying, Animating and Analyzing Biomolecular Systems
- Post Simulation Analysis
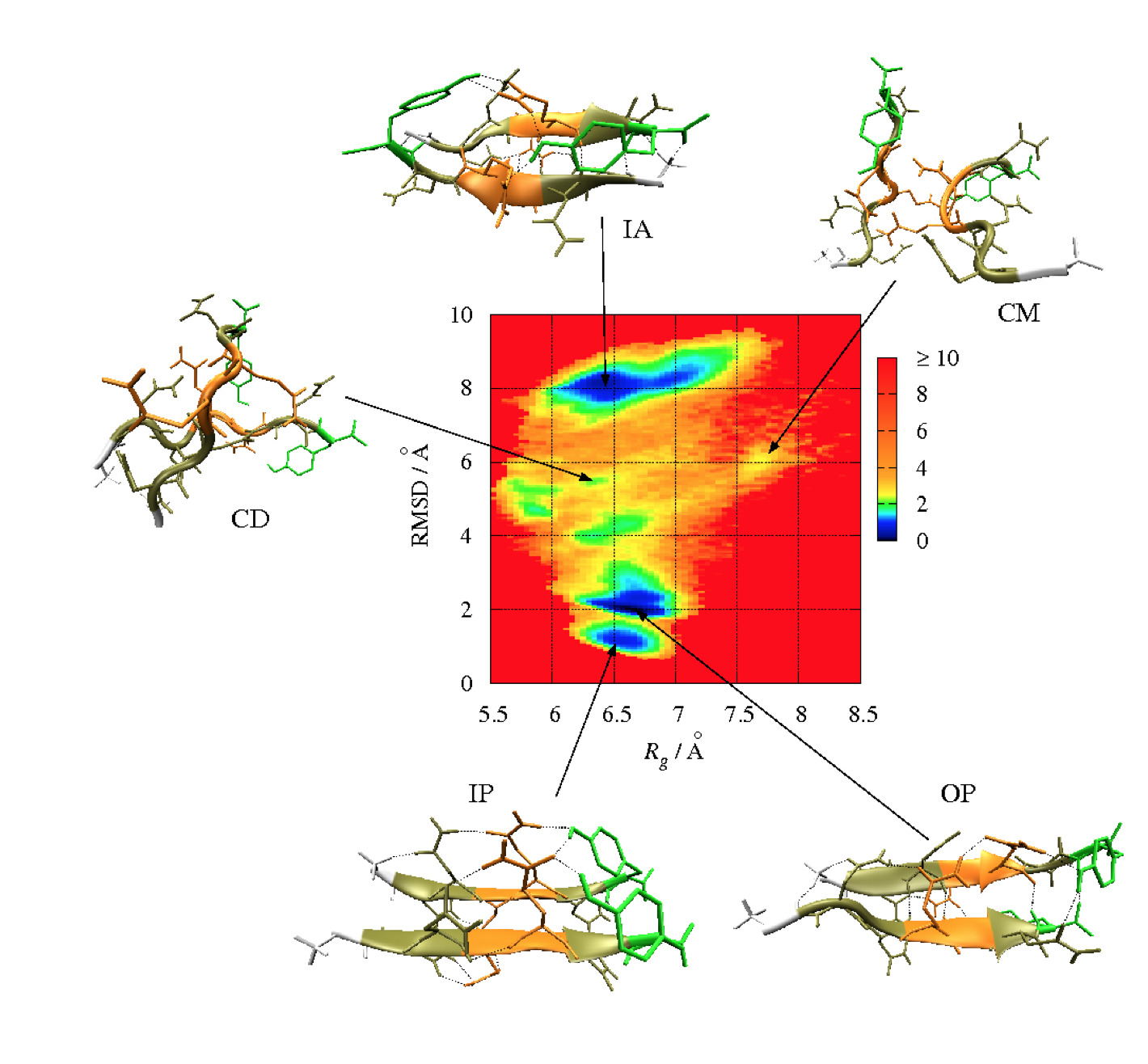
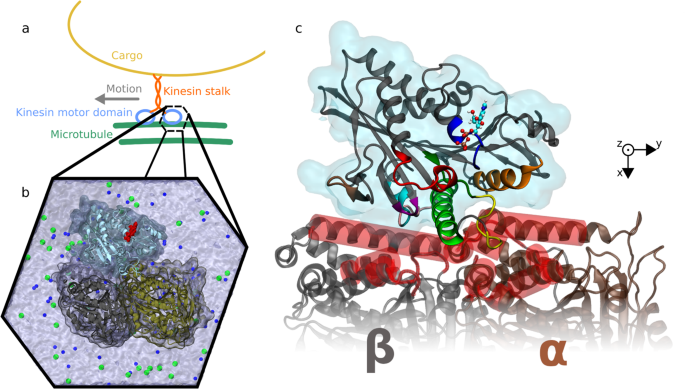
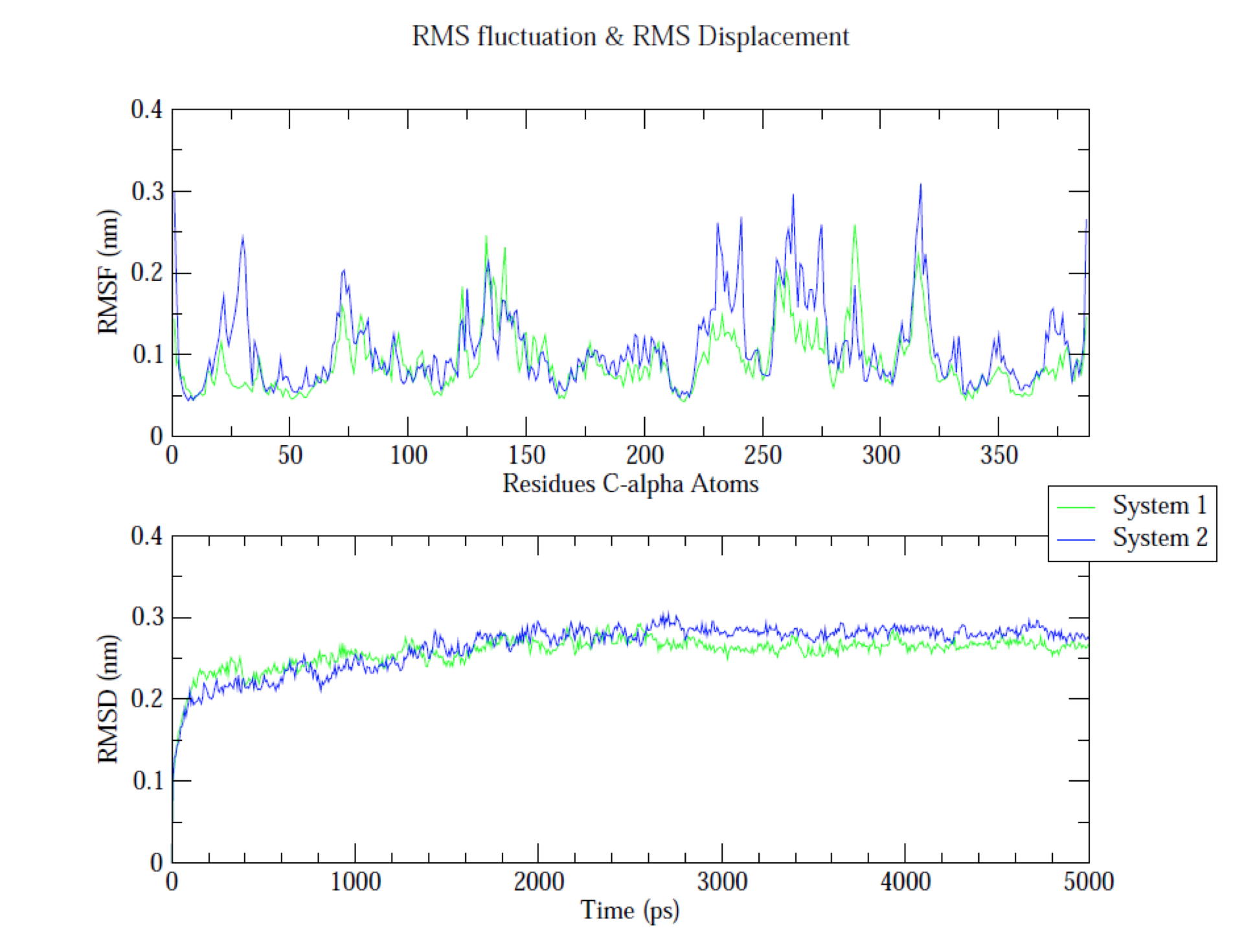
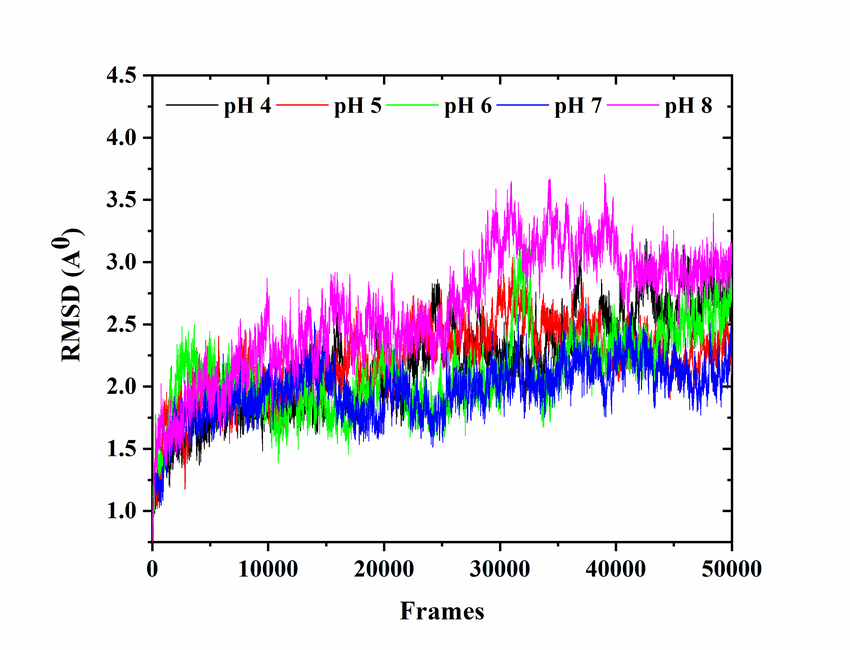
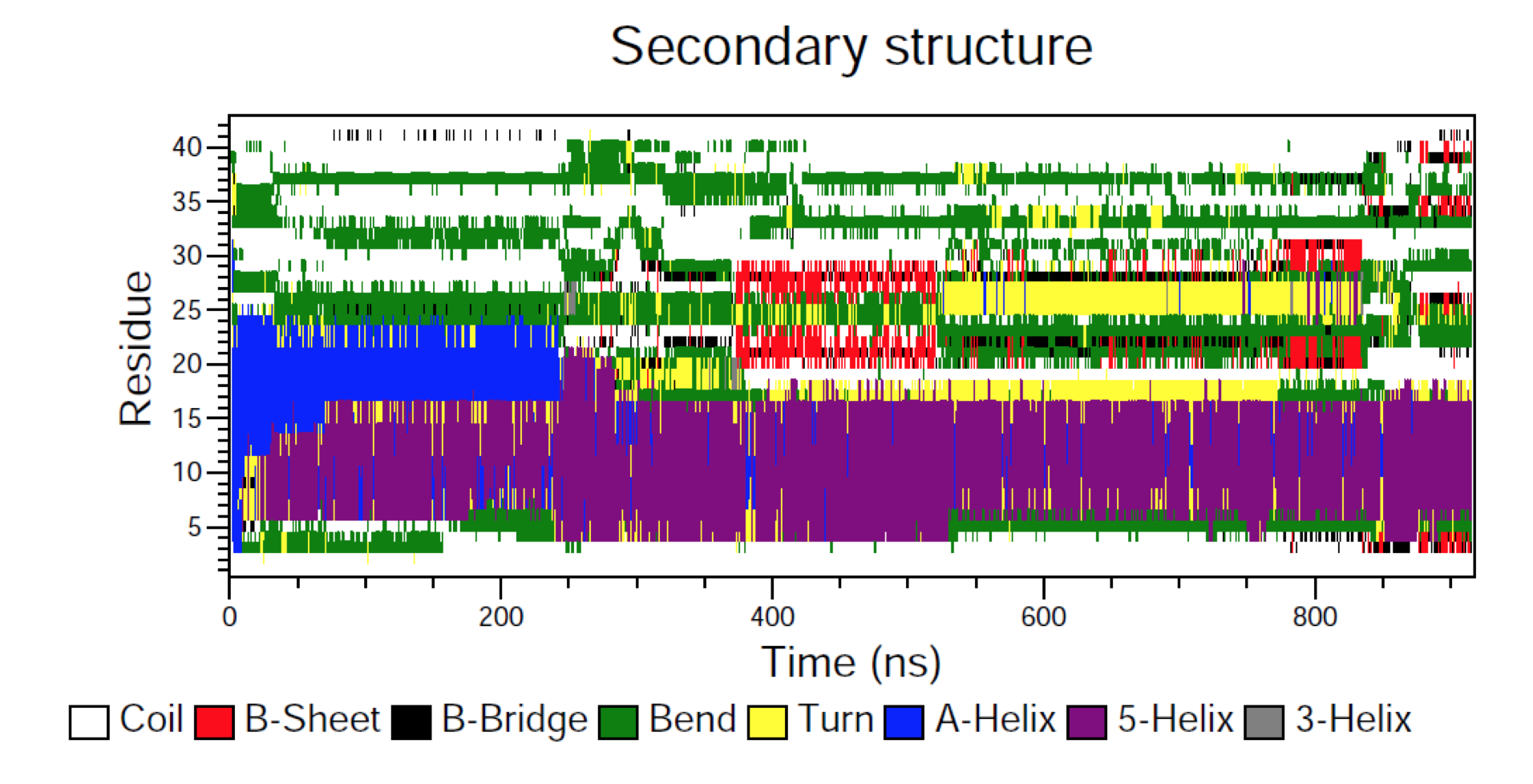
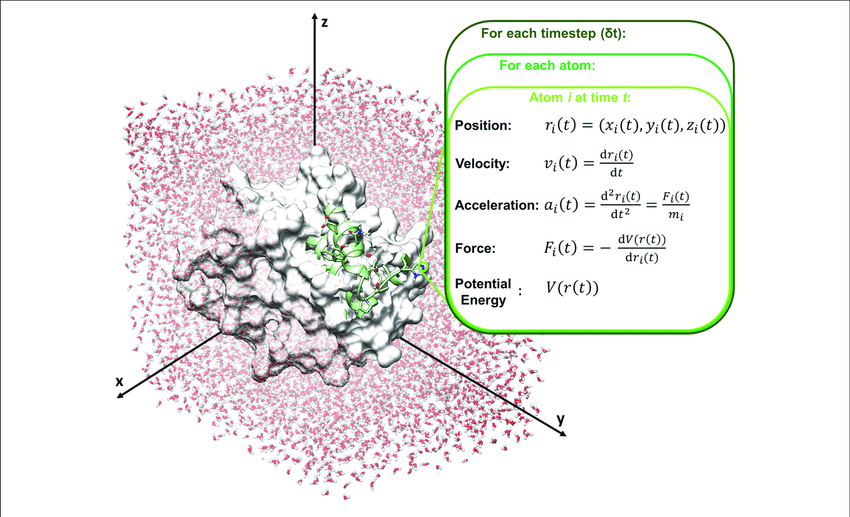
Molecular dynamics (MD) is a computer simulation method for analyzing the physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamic "evolution" of the system. In the most common version, the trajectories of atoms and molecules are determined by numerically solving Newton's equations of motion for a system of interacting particles, where forces between the particles and their potential energies are often calculated using interatomic potentials or molecular mechanics force fields. MD simulations are nowadays routinely applied to macromolecular systems of biological and pharmaceutical interest. MD simulation analysis is one of the essential steps while designing novel drugs using computational approaches.
Atomistic computer simulations of macromolecular (for example, protein) receptors and their associated small-molecule ligands play an essential role in drug discovery. The static models produced by NMR, X-ray crystallography, and 3D structure prediction provide valuable insights into macromolecular structure, but molecular recognition and drug binding are very dynamic processes. When a small molecule like a drug (for example, a ligand) approaches its target (for example, a receptor) in solution, it encounters not a single, frozen structure, but rather a macromolecule in constant motion. Simulations have proven valuable in breaking down the functional mechanisms of proteins and other biomolecules, in uncovering the structural basis for disease, and in the design and optimization of small molecules, peptides, and proteins. These simulations can capture a wide variety of important biomolecular processes, including conformational change, ligand binding, and protein folding, revealing the positions of all the atoms at femtosecond temporal resolution. Drug discovery provides a particularly interesting example of an area in which simulations can drive experiments. The future of computer-aided drug design is promising and hence molecular dynamics simulations play an increasingly important role in the development of novel pharmacological therapeutics.
Protein Structure Preparation
In order to perform Molecular Dynamics Simulations, first a protein structure should be pre-processed so that all the unwanted chains, ligands and unnecessary structural features should be removed so the next phases of molecular dynamics simulation will be executed correctly. You can prepare the protein structure for simulation using various protein structure visualization tools or through the Linux terminal.
Topology File Construction for Simulation
The next phase in molecular dynamic simulation after preprocessing of protein structure, is creating a topology file for the protein structure. If we are performing the molecular dynamics simulation through GROMACS, the GROMACS program pdb2gmx generates a topology for the input coordinate file. Several formats are supported for that coordinate file, but PDB is the most commonly-used format (hence the name pdb2gmx).
Defining a Solvent box for simulation
The next phase in the molecular dynamic simulation after pre-processing of protein structure and construction of topology file is to put the protein structure in a particular solvent box so that it could be simulated in the real world environment. The GROMACS program ‘editconf’ generates a box around the selected protein structure and edits the configuration of the protein structure file.
Solvation (Adding Water Molecules in Solvent Box)
After creating a solvent box for simulation, the next phase in molecular dynamic simulation is to perform solvation. The GROMACS program ‘solvate’ can be used to perform the solvation within the protein structure and to solvate the solute configuration.
Generating Input Run File (Replacement of Water Molecule with Ions)
The next phase in MD simulation analysis after solvation is to replace water molecules with ions, but before that an input run file should be created so that input run file can be used to create ions. The GROMACS program ‘grompp’ is used to make a run input file. The input run file basically consists of parameters that describe the atoms of the protein structure at atomic level.
Replacement of Water Molecules with Ions
The next phase in MD simulation analysis after generating the input run file is to replace water molecules with ions. The GROMACS program ‘genion’ is utilized to randomly replace solvent molecules with mono-atomic ions. The group of solvent molecules should be continuous and all molecules should have the same number of atoms.
Energy Minimization
Once you’ve created input files for running simulation, defined a solvant box, replaced the water molecules with ions, the next step is to minimize the energy of the protein structure and fixing the structure of the protein to make sure that there is no inappropriate geometry left in the protein structure, so that further MD simulation analysis can be performed smoothly. Energy minimization is essential to determining the proper molecular arrangement in space since the drawn chemical structures are not energetically favorable.
Visualization and Analysis of Minimized Structure
Once the simulation has been performed on the protein structure using the ‘mdrun’ command, the next step is to minimize the energy of the protein structure followed by its visualization and analysis. The GROMACS program ‘gmx energy’ is utilized for minimizing the energy of the protein structure while ‘gmx grace’ is utilized to create a visualization and analysis of the energy minimization plot in 2D.
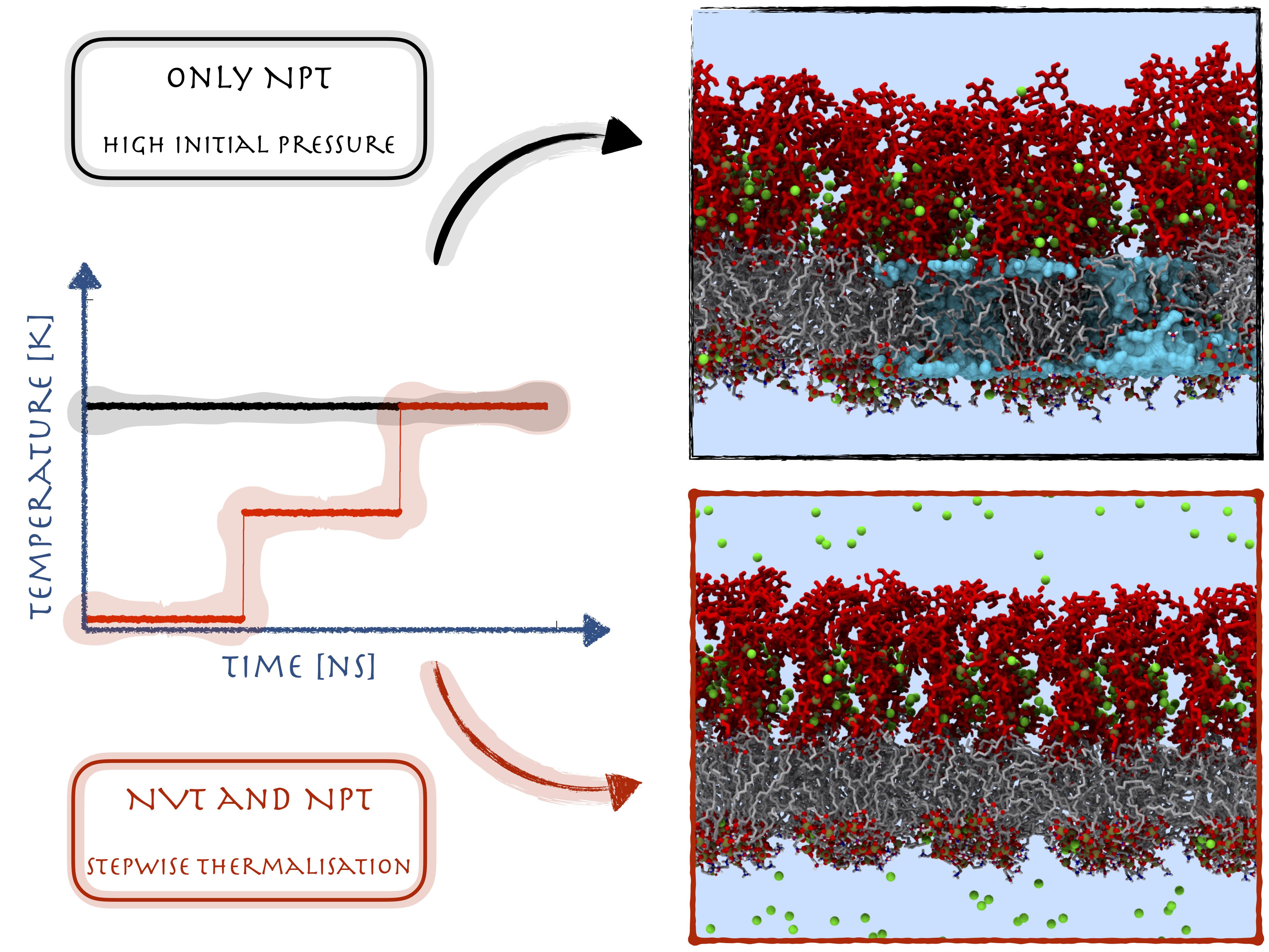
Equilibration of Protein Structure NVT ENSEMBLE (Phase 1)
Once you’ve minimized the energy of the protein structure on which further molecular dynamic simulation has to be done, the next step is to equilibrate the protein structure to optimize the structure and bring the solvant at the temperature where we perform the simulation analysis. And once that optimal temperature has been achieved, the next step will be to apply pressure to create a density within the protein structure so we can perform the simulation analysis at that region. Equilibration of proteins has to be performed in two consecutive phases. The first phase of NVT ENSEMBLE involves temperature stabilization of the entire system.
Equilibration of Protein Structure NPT ENSEMBLE (Phase 2)
Once you’ve brought the solvant the optimal temperature in the 1st phase of NVT ENSEMBLE, the next step is to apply pressure on the protein structure to bring the solvent in optimal density where further simulation can be performed on the structure. The constant-temperature, constant-pressure ensemble (NPT) allows control over both the temperature and pressure.
Executing Simulation Analysis
The GROMACS program ‘mdrun’ is the main computational chemistry engine within GROMACS. It is utilized to perform Stochastic Dynamics, Energy Minimization, test particle insertion or calculation/recalculation of energies. Hence through the mdrun program we run the MD simulation on the assembly structure of the protein and get simulation results.
Tools for Molecular Dynamics Simulations
Following are the tools that we use for Molecular Dynamics Simulation:
- GROMACS
- Desmond
- CHARMM
- AMBER
- NAMD
CHARMM
CHARMM (Chemistry at HARvard Molecular Mechanics) is a highly versatile and widely used molecular simulation program. It has been developed with a primary focus on molecules of biological interest, including proteins, peptides, lipids, nucleic acids, carbohydrates, and small molecule ligands, as they occur in solution, crystals, and membrane environments. The program provides a large suite of computational tools that include numerous conformational and path sampling methods, free energy estimators, molecular minimization, dynamics, and analysis techniques, and model-building capabilities.
GROMACS
GROMACS is a molecular dynamics package mainly designed for simulations of proteins, lipids, and nucleic acids. GROMACS is one of the fastest and most popular software packages available and can run on central processing units (CPUs) and graphics processing units (GPUs).
Desmond
Desmond is a software package developed to perform high-speed molecular dynamics simulations of biological systems on conventional computer clusters. The code uses novel parallel algorithms and numerical methods to achieve high performance on platforms containing multiple processors, but may also be executed on a single computer.
AMBER
AMBER (Assisted Model Building with Energy Refinement) is a family of force fields for molecular dynamics of biomolecules. AMBER is also the name for the molecular dynamics software package that simulates these force fields. The AMBER software suite provides a set of programs to apply the AMBER force field to simulations of biomolecules.
NAMD
NAMD (Not Another Molecular Dynamics Program) is a parallel molecular dynamics code designed for high-performance simulation of large biomolecular systems. NAMD uses the popular molecular graphics program VMD for simulation setup and trajectory analysis, but is also file-compatible with AMBER, CHARMM, and X-PLOR. NAMD is distributed free of charge with source code. It is noted for its parallel efficiency and is often used to simulate large systems (millions of atoms).
Post Simulation Analysis
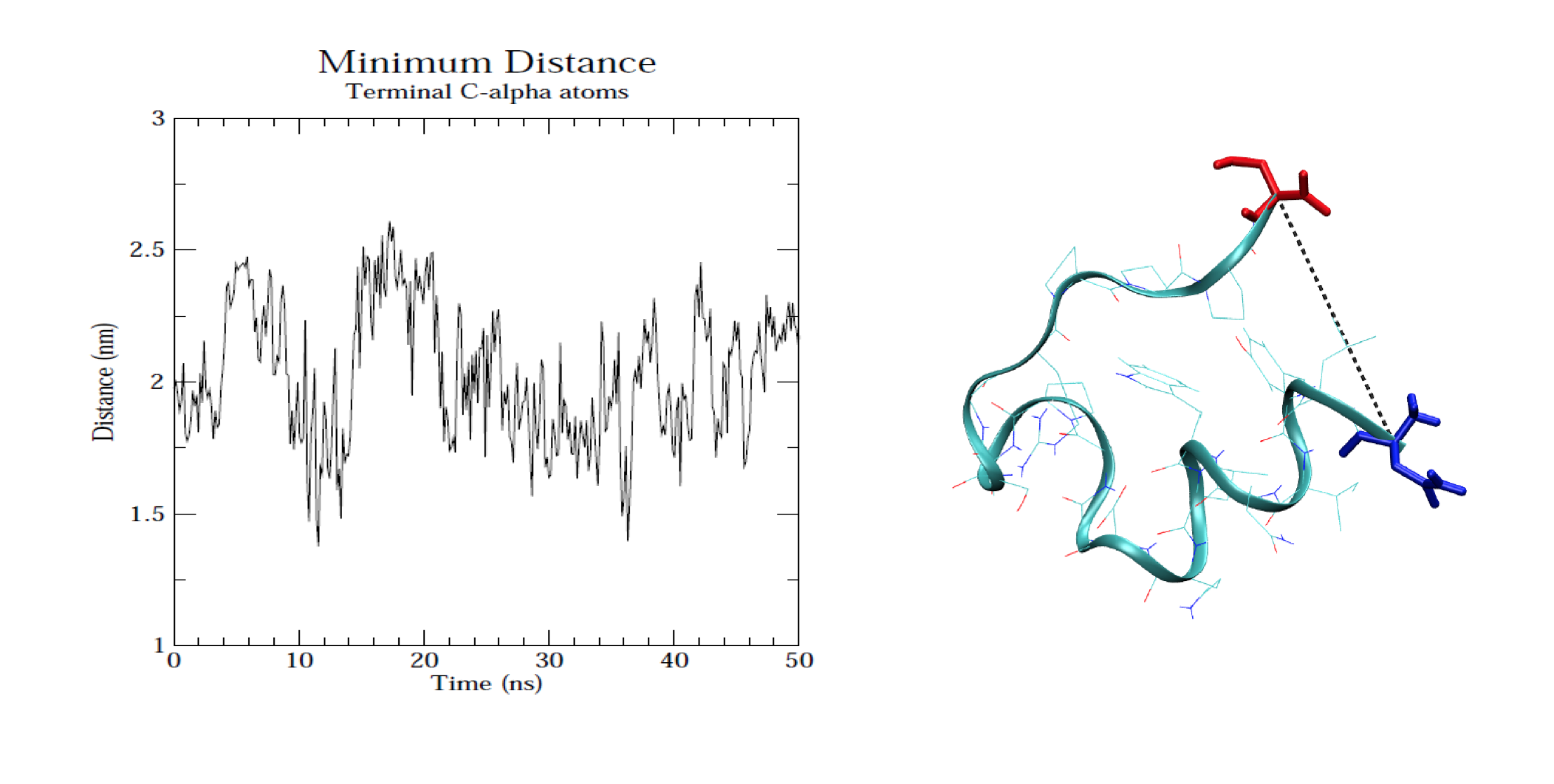
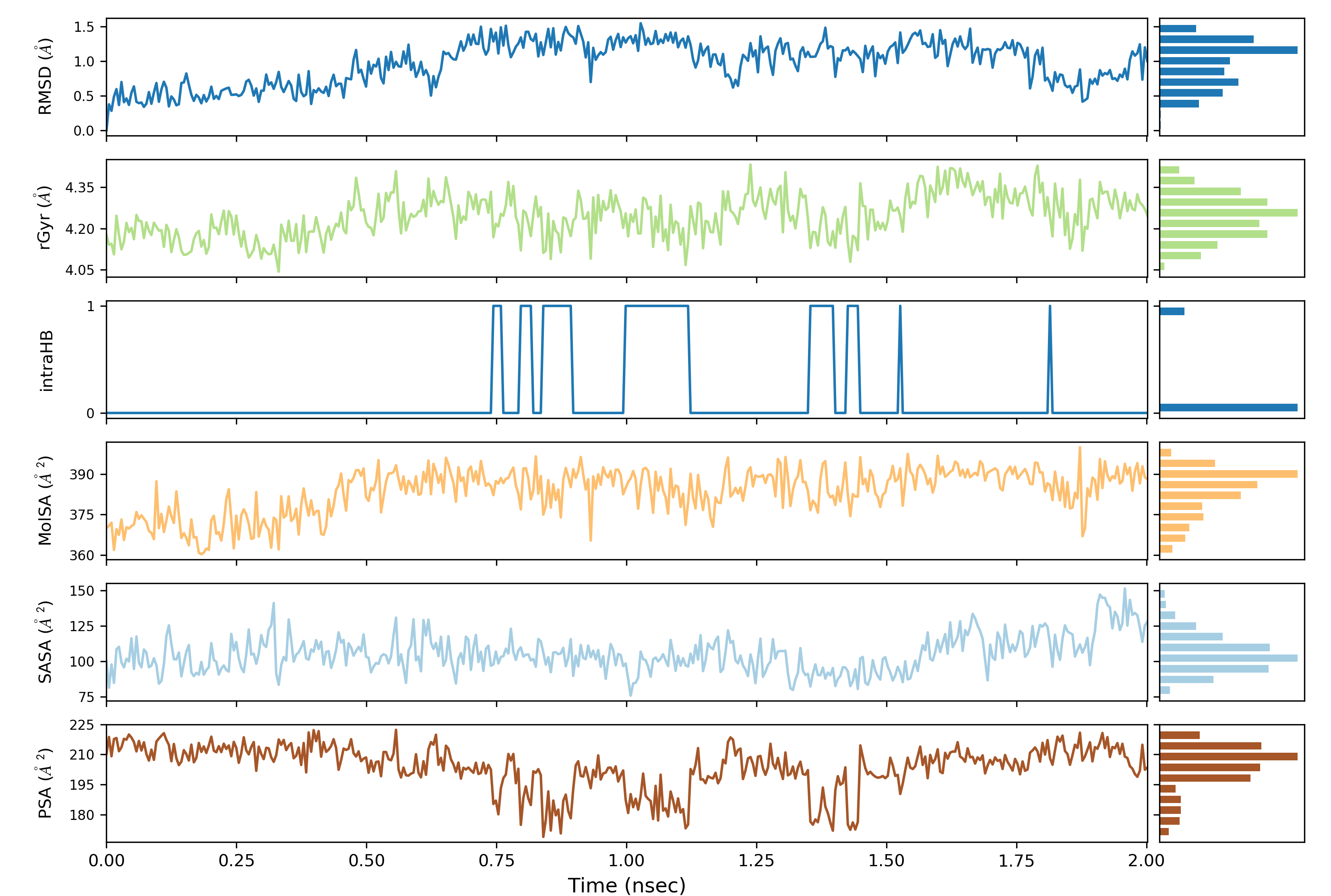
In order to analyze the molecular dynamics simulation results first we calculate RMSD (root-mean-square-deviation) to measure the average distance between a group of atoms (backbone atoms of proteins). Then we calculate RMSF (root-mean-square fluctuation) that measures the average deviation of a particle (a protein residue) over time from a reference position (typically the time-averaged position of the particle). After that principal component analysis (PCA) is performed which converts a set of correlated observations (atoms movement in protein) to a set of principal components which are linearly independent. Then MMPBSA is performed and GROMACS tool g_mmpbsa is used for performing high-throughput MM-PBSA calculations. After that hydrogen bond analysis is performed to identify the number of hydrogen bonds. Then Umbrella sampling is performed to find out the delG value in the protein-ligand system. After that a contact matrix is plotted.
Post simulation analysis can be done through the following ways:
- RMSD (root-mean-square-deviation)
- RMSF (root-mean-square fluctuation)
- Hydrogen Bond Analysis (H-bonding plot with time, H-bonding details)
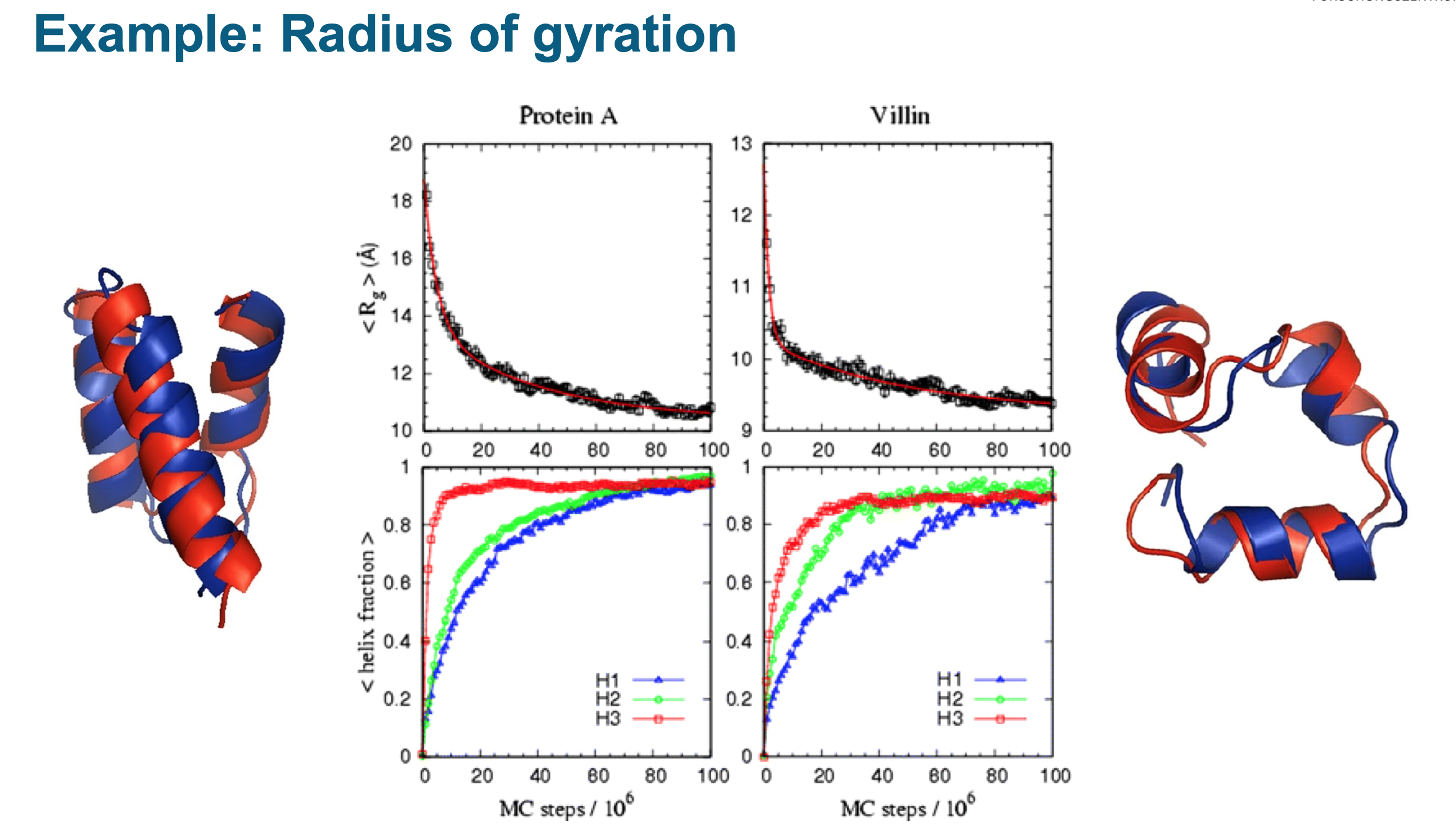
- Radius of Gyration (Rg)
- SASA (Solvent Accessible Surface Area)
- Contact Frequency Analysis or Contact Area Analysis
- Protein-ligand center of mass distance
- Principal Component Analysis (PCA)/ Dynamic Cross-Correlation Matrix Analysis (DCCM)
- Salt-Bridge Analysis
- MMGBSA/MMPBSA Binding Free Energy (with its decomposition)
Get in touch with us and we will start your analysis right away!
