BioCode is offering one-on-one live training for beginners who are struggling with learning bioinformatics or are stuck in their bioinformatics analysis.

Each and everything related to bioinformatics will be taught in this live training, followed by guidance on the analysis of your project. Our hands-on training will make sure that you learn every little thing about the theoretical and practical aspects of your bioinformatics analysis.
Training Schedule & Features
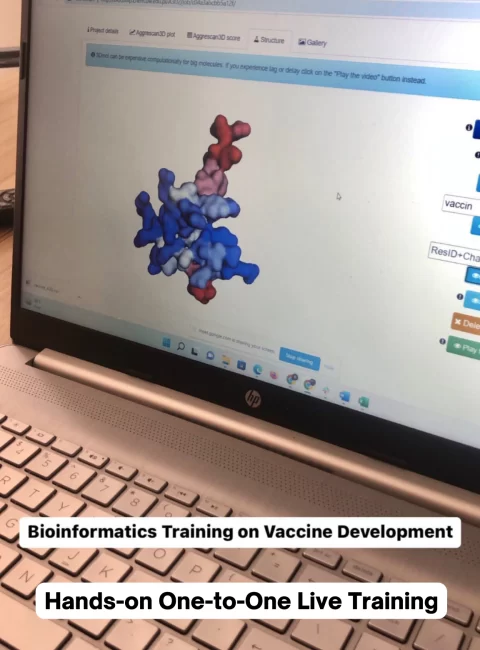
Features of training:
-Training is conducted 1-1 between you and the tutor
-Training starts immediately after the payment
-Beginners with no experience are encouraged to enroll
-Advanced level industrial skills will be taught
-Communication between you and the tutor
-Assistance in paper writing and publishing
-2 hours/per class, twice a week
-Classes: 8/16
-You will be added to our company Slack channel for communication
-Online meetings will be conducted through Google Meet
Multiple Training Categories (You can enroll in any)
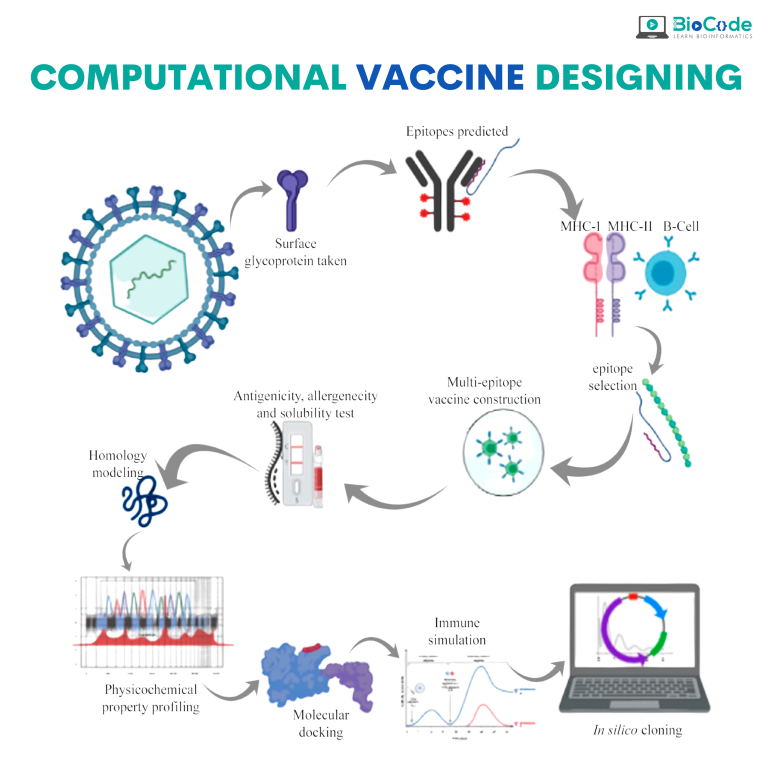
Category 1: Computational Vaccine Designing
Description: This session will focus on making the participants learn how to computationally develop a vaccine from end to start.
Learning Outcomes: Upon completion of this session, participants will be able to:
-
Retrieve the Proteome, Antigenicity, and Location Prediction.
-
Predict and Evaluate Cytotoxic T-Lymphocytes (CTL) Epitopes.
-
Predict and Assess Helper T-Lymphocytes (HTL) & Linear B-Lymphocytes (LBL) Epitopes.
-
Map Vaccine Construct.
-
Estimate Population Coverage.
-
Predict & Analyze Primary & Secondary Structure.
-
Predict, Refine & Valide Tertiary Structure.
-
Screen B Cell Epitopes.
-
Perform Molecular Docking Between Vaccine & TLR4 receptor.
-
Perform Disulfide Engineering of Vaccine.
-
Perform Codon Optimization and In Silico Cloning.
-
Perform Molecular Dynamics & Immune Simulation.
Category 2: Computational Drug Designing
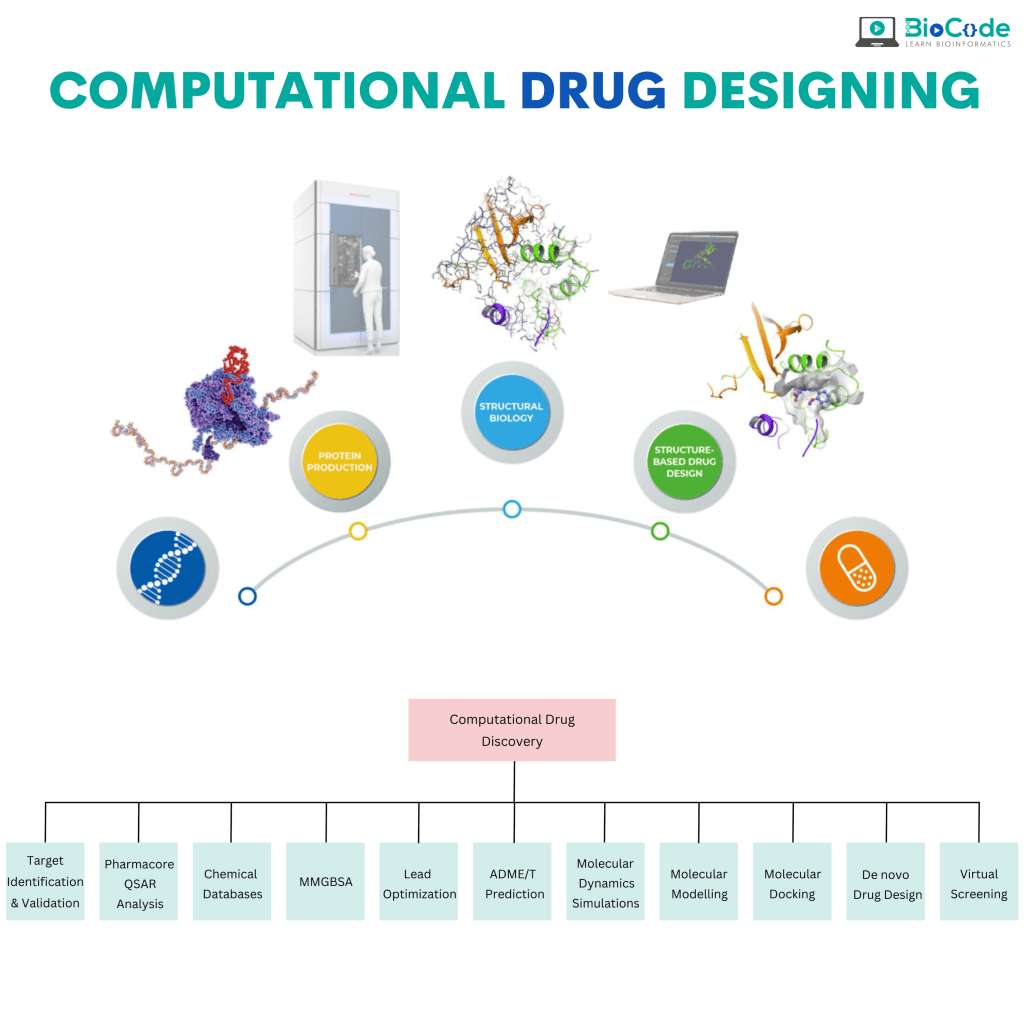
Description: This session will focus on making the participants learn how to computationally develop a drug from end to start.
Learning Outcomes: Upon completion of this session, participants will be able to:
-
Explain the Role of Bioinformatics in Drug Designing.
-
Perform Lead Identification.
-
Assess ADMET Properties.
-
Perform Protein 3D Structure Prediction.
-
Perform Molecular Docking.
-
Understand Reverse(Computational) Vaccinology & Immunoinformatics.
-
Perform Drug Repurposing.
Category 3: Biological Programming
Description: This session will make sure that every participant learns biological scripting from basics to advanced level.
Learning Outcomes: Upon completion of this session, participants will be able to:
Describe Python, BioPython, R, Linux & BioConductor.
Perform File Handling and Error Handling.
Perform Bioinformatics File Parsing and Writing.
Explain Loops and Functions.
Perform Phylogenetic Analysis in BioPython.
Perform Data Visualization using ggplot2 in R.
Write Customized Scripts in Linux.
Perform Advanced Biological Data Analysis Using Python and R.
NGS Analysis Using BioConductor Packages.

Category 4: Cancer Genomics
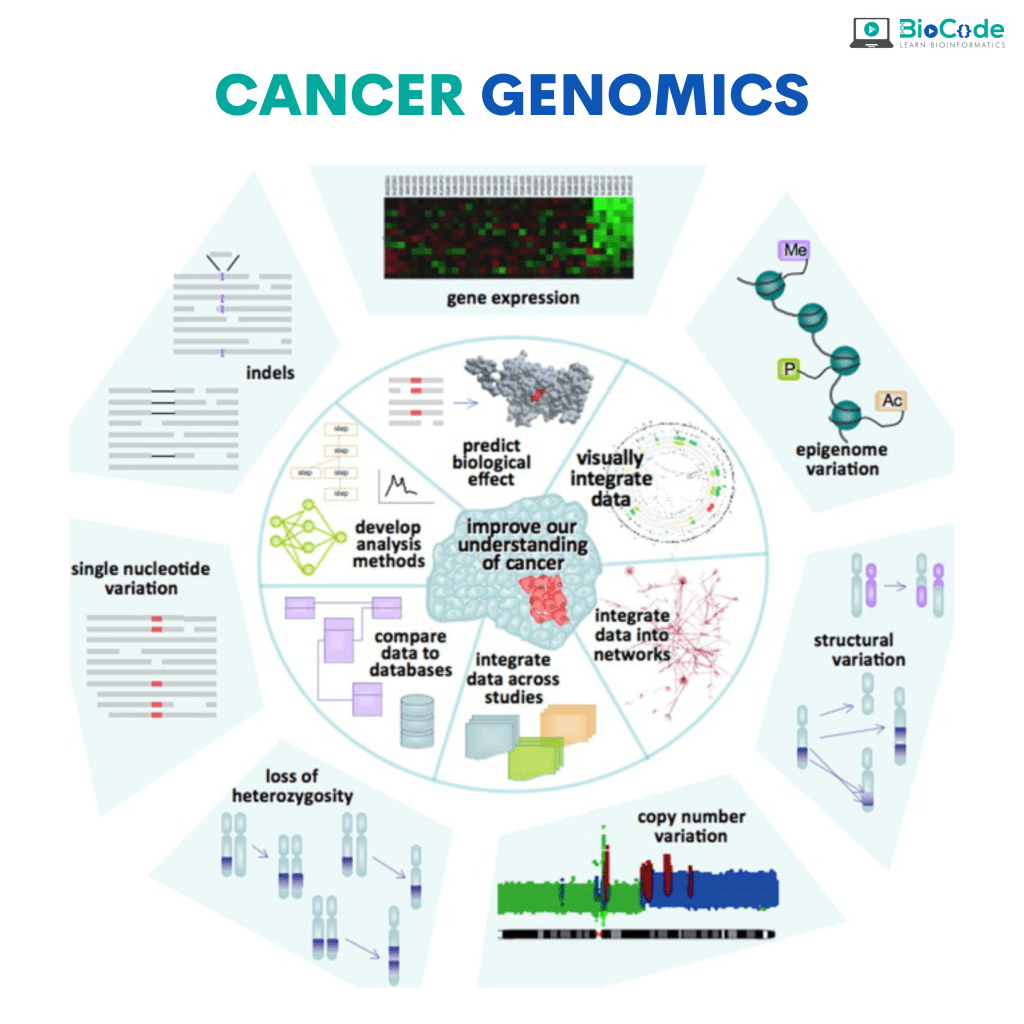
Description: This session will focus on making sure that the participants understand cancer genomics and various analysis necessary in cancer genomics.
Learning Outcomes: Upon completion of this session, participants will be able to:
Understand Cancer Genomics and Genomics.
Perform Analysis of Cancer Patients’ Biological Dataset
Identify Somatic and Germline Mutations in Cancer Patients.
Perform Genome-Wide Association Analysis.
Perform Quantitative Loci Analysis (eQTL).
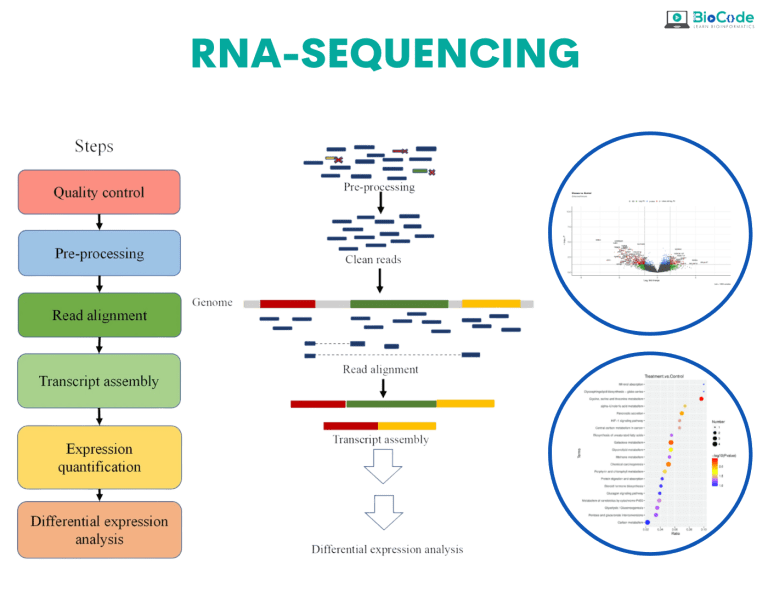
Category 5: RNA-Sequencing
Description: This session will focus on making sure that the participants understand RNA-sequencing and how the RNA-sequencing pipeline works.
Learning Outcomes: Upon completion of this session, participants will be able to:
Understand RNA-Sequencing and RNA-Sequencing Pipeline.
Retrieve Raw Dataset (Raw FastQ files).
Perform Quality Control and Trimming.
Perform Read Mapping/Alignment.
Discover Differentially Expressed Genes.
Perform Functional Enrichment Analysis.
Category 6: ATAC-Sequencing
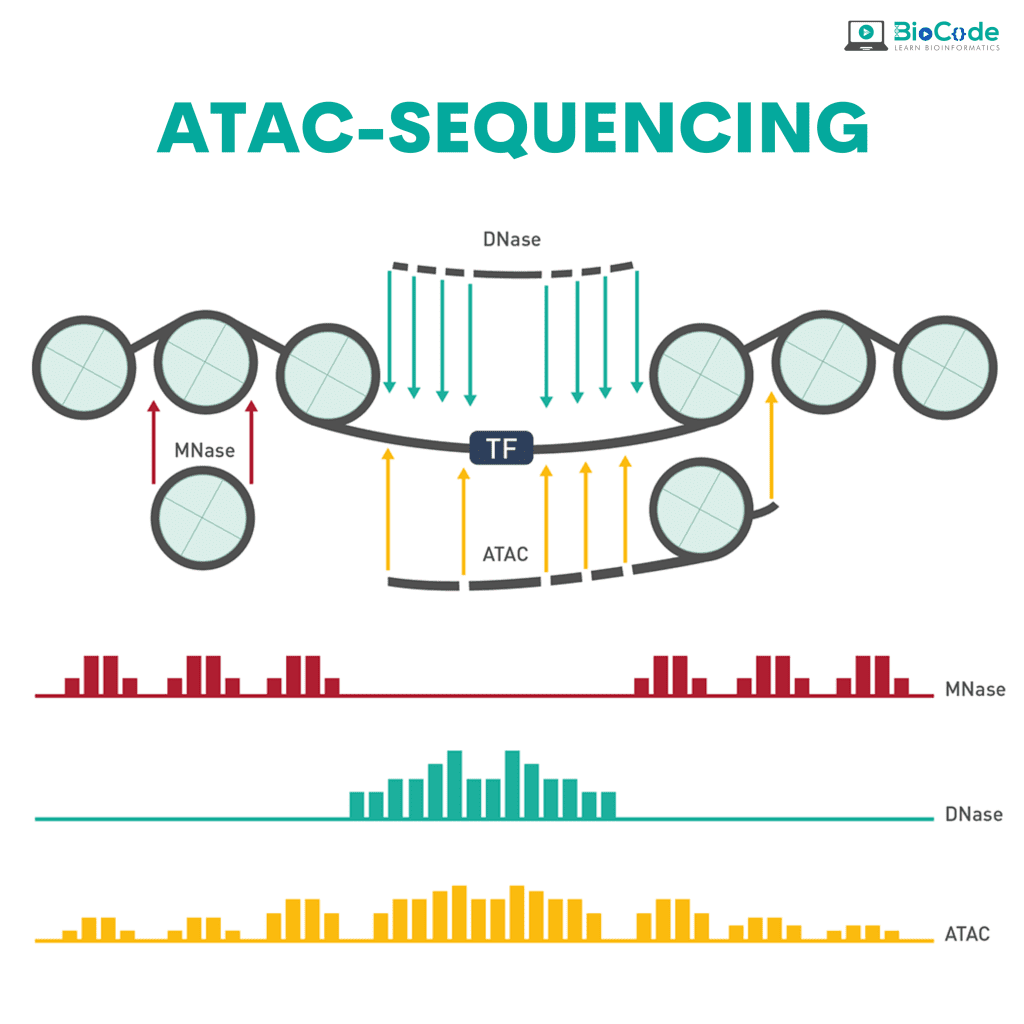
Description: This session will focus on making sure that the participants understand ATAC-sequencing and how the ATAC-sequencing pipeline works.
Learning Outcomes: Upon completion of this session, participants will be able to:
Understand ATAC-Sequencing and ATAC-Sequencing Pipeline.
Explain Pre-Analysis, Core Analysis, and Advanced Analysis in ATAC-Sequencing.
Retrieve Raw Dataset (Raw FastQ files).
Perform Quality Control and Trimming.
Perform Read Mapping/Alignment.
Perform Post-Alignment Processing.
Perform Peak Calling.
Perform Peak Differential Analysis and Annotation.
Perform Motif Enrichment Analysis and Footprinting Analysis.
Perform Nucleosome Positioning.
Integrate ATAC-Seq with ChIP-Seq and RNA-Seq.
Construct a Regulatory Network.
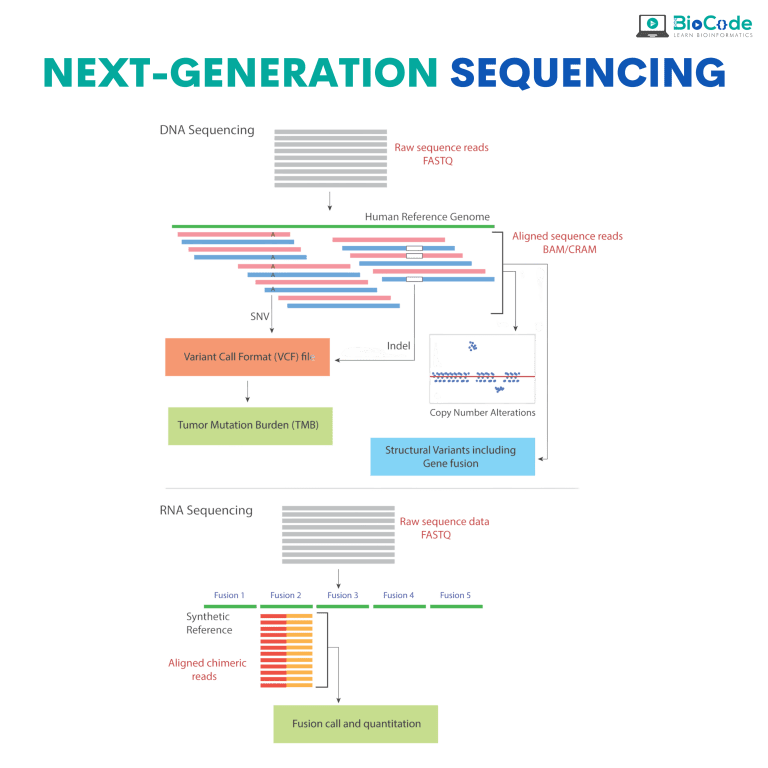
Category 7: Next-Generation Sequencing
Description: This session will focus on making sure that the participants learn about Next-Generation Sequencing (NGS) and NGS analysis pipelines.
Learning Outcomes: Upon completion of this session, participants will be able to:
Give an In-Depth Introduction to NGS and its Pipelines.
Explain Technologies and Machines Involved in NGS Analysis.
Perform Variant Calling to Identify Mutations in Raw Datasets.
Perform Differential Expression Analysis.
Identify Transcription Factors Binding Sites.
Identify Histone Modifications.
Category 8: Metagenomics
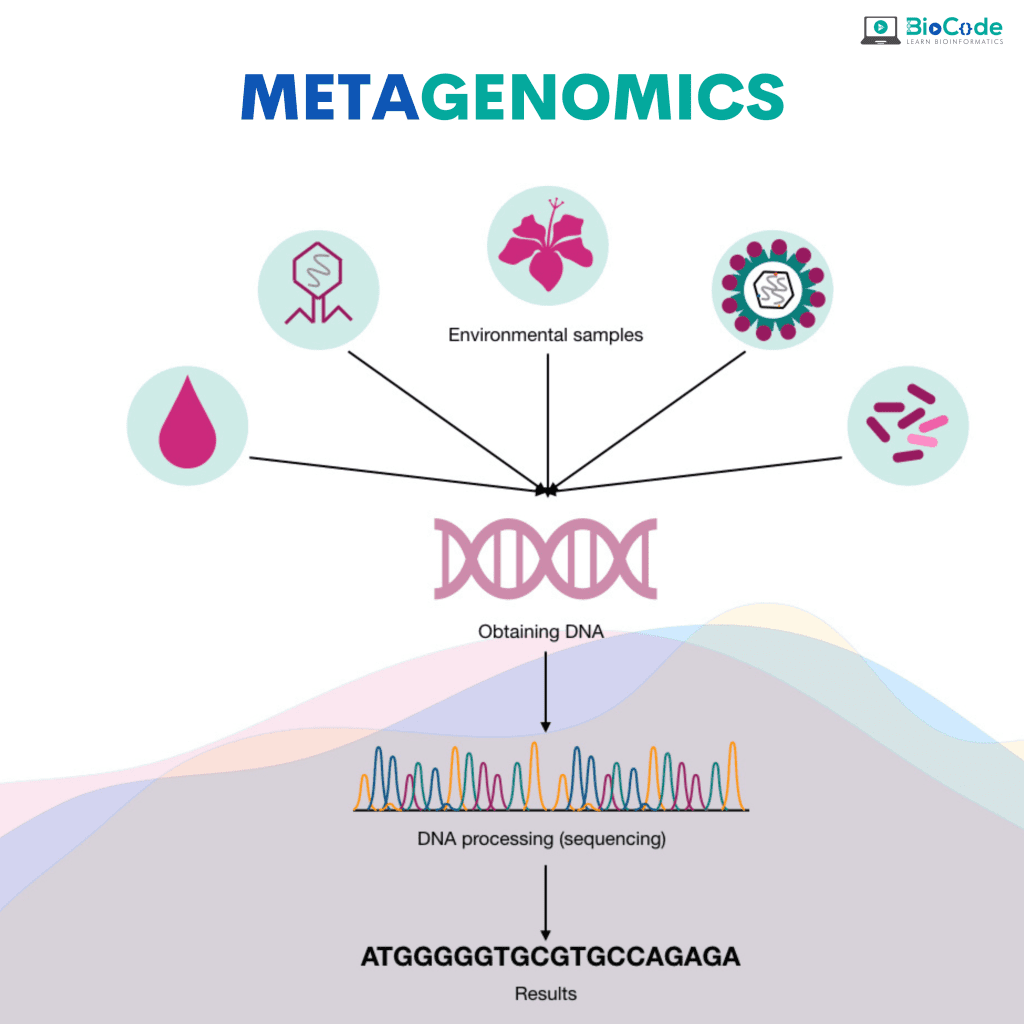
Description: This session will make sure that the participants learn about the underlying concepts of metagenomics.
Learning Outcomes: Upon completion of this session, participants will be able to:
Explain and Perform a Complete Metagenomics Pipeline.
Identify Species Present in Gut Microbiome and Soil Microbiome.
Get Started With Enrollment
BioCode gives you the opportunity with getting trained in industrial level advanced skills that can greatly enhance your Bioinformatics and Life Sciences career and land you your dream job in the field.
To enroll in the personal 1-1 training between you and the tutor for your desired categories mentioned above, please fill the form and our team will get in touch with you.
- Duration: 1 months
- Classes: 8
- Live Classes: 1-2 hours each, twice a week
- Session: Google Meet
- Communications: Slack
- Training level: from basics to advanced
- Experience required: basics of biology
