What is Single Cell Genomics?
The last decade has observed advancements in the fields of cellular biology, molecular biology and bioinformatics. These advances have led to the advent of many new techniques including the “Single Cell Genomics (SCG)”. The emergence of SCG is due to improvements made in molecular cloning and microfluidic, which have allowed us to understand complicated biological phenomena by enhancing the resolution to a level of single cell, thus opening up a new era. This new era of SCG allows us to quantify millions of genes from numerous ‘single cells’ simultaneously from a sample or specimen.
Technically, SCG is defined as an advanced technique that provides cellular level genetic information from a single-cell genome. This technique evolved as the omics approaches were bettered with time, which allowed increased throughput and accuracy in isolation of single cells. As the isolation approaches were flourished, the genomics with single cell sequencing technologies opened up new insights to link functionality and phenotype of cell with genotype. Thus, enabling us to know genetic heterogeneity present in individual due to difference in development of normal and diseased cell.
Here a simple question comes to our mind. We already have many advanced genetic techniques like PCR, immunofluorescence, single-cell PCR, in situ hybridization and many other, what makes SCG better than these? The answer is simple, all of these techniques are based on molecular markers which are limited in number. This limitation is overcome by SCG, as it does not rely on molecular markers, it is a complete analysis of all the genes present in a single isolated cell.
Procedure of SCG technique
This technique involves steps which allow us to do genomic analysis of single cells. The steps involved in SCG includes:
Single Cell Isolation
DNA isolation from single cell
Amplification of single genome
Analysis of single genome
Single Cell Isolation:
As the technique is named as single cell genomics (SCG), so the first step is to isolate single cells for analysis. There are different principles used for cell isolation and on the basis of these we can classify cell isolation techniques into two groups. The first group of techniques the principle of isolation is based on physical properties like size, density, charge and physical deformation. These isolation techniques include microchip-based platforms and density gradient centrifugation. The advantage for these techniques is that the cells can be isolated without any need of labelling. In second group of techniques, the principle is based on biological properties and characteristics of cells. These techniques include affinity-based cell sorting i.e., magnetic-activated and fluorescence activated cell sorting. All the methods have certain limitations. Some of the isolation techniques are as follows:
Membrane Filtration: In this cell isolation method, a membrane filter having pores is used. The cells are isolated on the base of size. The membranes consist of all shapes and sizes of pores, it depends upon the size and shape of cell we want to trap on porous membrane. This method is very laborious, time-consuming and error-prone. These limitations are overcome by using other cell isolation techniques.
Fluorescently activated Cell sorter: It is a modern and easy cell isolation technique. It is a special type of flow cytometry which has ability to sort different cells on the characteristics like size, granular properties and fluorescence. This technique allows the analysis of single cells both quantitative and qualitative simultaneously.
For this method, cell suspension is prepared with fluorophore conjugated monoclonal antibodies labelled target cell. When they are exposed to laser light, the detectors identify the target cells and their particular properties. Then, the device gave the charge on target cell, either a positive charge or a negative charge. This charged particle is then collected by electrostatic deflection system in tubes.
Magnetic-Activated Cell sorting (MACs): The cells are isolated by this technique on the basis of their surface antigen. It allows the separation of pure cell population at about 90%. It uses magnetic beads bounded enzymes, antibodies or lectins. These molecules will associate with specific type of protein present on the target cell. After their association, when these magnetically labelled cells are placed under the outside magnetic field, then, they are polarized and are collected by elution. While all the unpolarized cells are washed out. This technique is faster and more efficient as compared with fluorescently labelled cell sorting. But on the other hand, it is quite expensive.
Laser Capture Microdissection (LCM): It is considered to be a most efficient and accurate cell sorting method where microscopic slide tissue samples are used. LCM has two types i.e., ultraviolet or infrared. The system of LCM is composed of an ultraviolet or infrared laser diode, inverted microscope, a laser control unit, CCD camera, microscope stage which is controlled by joystick supplemented with vacuum chuck for immobilization of slide and color monitor.
In this method, the target cells containing tissue samples are present on slide which can be viewed under microscope. Afterwards, these tissue sample slides are exposed to laser pulse for short duration. This results in liquification of thin and transparent thermoplastic film from the cells of interest. The film, which is liquified, then fuse with target cells. Then, as the thermoplastic film is removed which now have the cells of interest. In addition to this, these isolated cells can be centrifuged with appropriate buffer solution for analysis. Overall, LCM is an accurate, fast and efficient method to isolate pure populations of cells.
Also, it is important to know that, while we isolate our desired cells by using LCM, the adjacent cells present in the tissue samples are not destroyed or affected. So, if the analysis required isolation of the adjacent cells, then, it can be easily done. But the limitation faced in overall LCM is the high technical skills are required for accurate and precise isolation of cells. As we need a close microscopic inspection of the cells for their required characteristics, therefore, without a proper trained cytologist or pathologist, proper identification of cells cannot take place.
Manual Cell Picking (MCP): It is also known as the micromanipulation. MCP is a very important technique which allow us to isolate pure cells from cultures or embryos. This method contains movable micropipettes with an inverted microscope. Microscope is use to observe cells and then they are photographed, allowing to be isolated accurately. This is a very simple, easy and precise technique for single cell isolation. But as it is a manual method, so the output is limited or low and high level of professional skills are required.
Microfluidics (MF): It is considered as one of the powerful techniques which enables us to understand the intrinsic complexity present in cellular system by means of a very small amount of sample. Microfluidic cell sorting can be divided into four types:
Cell affinity chromatography based microfluidic isolation
Immunomagnetic beads based microfluidic isolation
Physical features based microfluidic isolation
Dielectric properties of cells based microfluidic separation
In these above-mentioned methods, most frequently used is the cell affinity chromatography based microfluidic isolation. This method contains specific types of interactions as present between antibody and antigen or ligand and the receptor. In this first step, chip’s microchannel are altered or modified by using specific antibodies. These antibodies have affinity to bind with the cell surface antigen of the target cell. So, as the antigen antibody interaction takes place, the cell becomes immobilized on the chip. The other cells are eluted by using a buffer. In the end, immobilized cells are eluted for downstream analysis by using a different buffer. These affinity-based methods are rapid, efficient and accurate due to binding by specific recognition. Presently, MF is being used in conjunction with techniques like MACS, filtration etc.
DNA Isolation from Single Cell:
After single cell isolation, the DNA extraction takes place. But firstly, the cell undergoes mild cell lysis and then DNA is extracted by using DNA extraction kits. Mild lysis of cell is done by using lysis buffer and it is done to remove membrane of single-cell. This allows compartmentalized DNA to be released which is then extracted by DNA extraction kits. High quality DNA is necessary for analysis, as broken or degraded DNA will have incomplete genome information. Also, such type of DNA cannot be evenly amplified.
Amplification of single genome:
After DNA isolation, it is subjected to DNA amplification, which allow us to amplify trace quantity of DNA from single-cell into large amount. Nowadays, two types of methods are used which are isothermal techniques and hybrid techniques. In hybrid techniques, amplification of single cell genome is done by modifying polymerase chain reaction (PCR) i.e., primer extension pre-amplification PCR, linker adaptor PCR etc. But all these methods have many limitations like amplification bias, low coverage and dropout of alleles.
On the other hand, isothermal methods include multiple displacement amplification (MDA) and multiple annealing and looping–based amplification cycle (MALBAC). These methods result in amplification at exponential rate and nonspecific priming. It has been showed by scientists that MDA has a better coverage of genome as compared to MALBAC, therefore, MAL has high detection rate per nucleotide, on the other hand MALBAC has shown to be useful in detecting copy number variations. Overall, these methods have shown less false-positive rates but more false-positive variance in labs. Recently a new method has been devised named as linear amplification via transposon insertion (LIANTI) which has proved to produce less bias and error.
Analysis of single genome:
The main aim of all the steps mentioned above is to make single cell enable for analysis, so that this analysis can be used for various application. For analysis, whole genome amplification (WGA) is pre-requisite. After WGA, the products of single cell are observed and examined. This is done by targeting specific regions of genome which are considered to have higher biological input and also this decreases the probability of false-positive results. This target specific amplification of genome has high and uniform coverage.
Analysis of the whole genome can be done by using sequencing techniques. Single cell exome sequencing allows us to examine coding regions and to identify variants in genome which are cell-specific. But in this technique, as the region of genome is increased for analysis, the probability of getting false-positive results also increases. Besides exome sequencing, whole genome sequencing (WGS) can also be used. Whole genome sequencing enables to identify variations present in non-coding region as well. But WGS is quite expensive and almost 30 times more costly than exome sequencing.
NGS is advanced and high throughput sequencing method. It has higher resolution and precision as compared to microarray methods. NGS includes illumine sequencing, SOLiD sequencing and Roche 454. These techniques have greater accuracy in calling variants in single cell. All these platforms of NGS are used to screed amplified whole single cell genome for any type of alteration and variations.
One of the major challenges faced in single cell genome sequencing is the interpretation of SCG data. Sometimes it becomes difficult to distinguish between noise and technical fault. This can lead to false-positive results. Therefore, in order to overcome this, it is quite important to compare the variant allele with the bulk population, so that selection bias can be avoided. Also, while calling the single cell variants, it should be kept in mind that various errors can occur during WGA and analysis tools. No doubt, all the techniques are refined day by day to overcome noise, but while identifying variations all the imbalances of alleles and mutations during amplification must be noted. The following measures can be taken to avoid false positive results:
NGS is advanced and high throughput sequencing method. It has higher resolution and precision as compared to microarray methods. NGS includes illumine sequencing, SOLiD sequencing and Roche 454. These techniques have greater accuracy in calling variants in single cell. All these platforms of NGS are used to screed amplified whole single cell genome for any type of alteration and variations.
One of the major challenges faced in single cell genome sequencing is the interpretation of SCG data. Sometimes it becomes difficult to distinguish between noise and technical fault. This can lead to false-positive results. Therefore, in order to overcome this, it is quite important to compare the variant allele with the bulk population, so that selection bias can be avoided. Also, while calling the single cell variants, it should be kept in mind that various errors can occur during WGA and analysis tools. No doubt, all the techniques are refined day by day to overcome noise, but while identifying variations all the imbalances of alleles and mutations during amplification must be noted. The following measures can be taken to avoid false positive results:
For variant calling bulk data should be used.
Only those variations must be considered positive which are present in two to three single cells at the same location.
Variant calling algorithms are necessity as they can overcome allelic imbalances by taking technical noise into account.
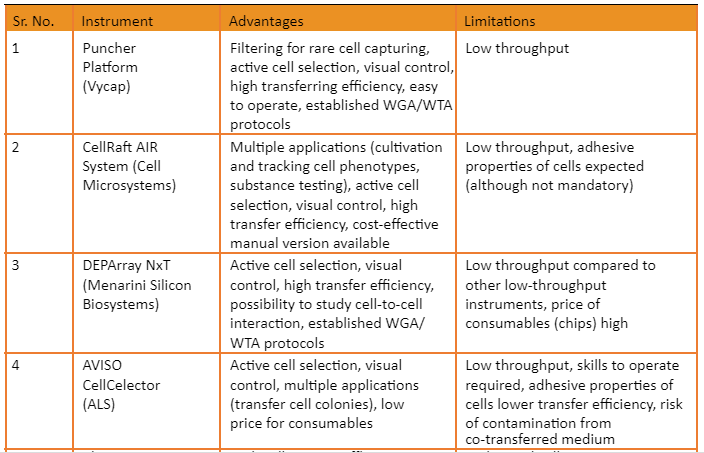
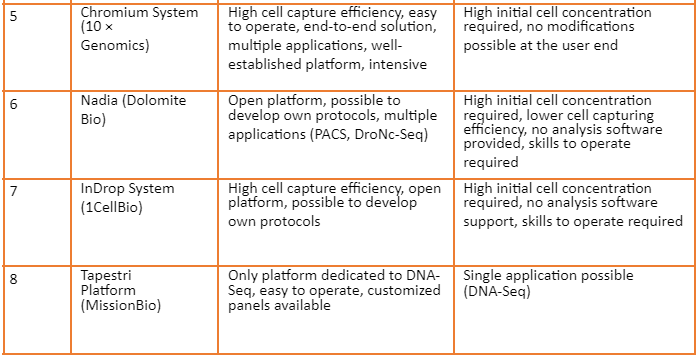
Overview of commercially available platforms is given by the following table:
Applications of SCG
SCG has grown rapidly in past few years. Its applications are present in almost every field of biology which includes immunology, cancer biology, microbiology, neurobiology and tissue mosaicism. Some of the important applications detail is given below:
In cancer Biology
As the cancer is caused due to uncontrolled division of undifferentiated cells, so SCG has ability to isolate these different blends and types of cells. This helps in targeted therapy of cancer, as SCG gives an insight on heterogeneity and gene modulation present in cell. All the stages of cancer i.e., tumor initiation, its maintenance and finally the evolution can be detected by genetic variation in single cell. SCG also help in identification of rare type of tumor cells. Thus, SCG is helpful in diagnostic of cancer, monitoring its growth and directing the targeted therapy according to cell type.
In Tissue Mosaicism
It was widely accepted theory that all the normal cells consist of same types of genes. This accepted principle was challenged by SCG by confirming the presence of mosaicism in genes. This genetic mosaicism is present is normal cells of an individual during his normal growth. SCG has confirmed that variations present in these normal cells are at great genomic tenacity.
In diagnostics
CSG is quite helpful in distinguishing normal and diseased cells. It is a very powerful tool to analyze the variations that are associated with a disease condition which can later on be cured by effective treatment.
In Immunology
The immune system consists of cells involved in innate and adaptive immune responses. SCG is useful in studying different types of immune cells which allow us to observe different types of rare and intermediatory cell states that cannot be studied in bulk population.
In Microbiology
SCG is very useful and powerful in studying different microbial community research. SCG allow us to build evolutionary relationships among microbes by analysis of single cells. They also enable us to study microbial features of various microbes. Through SCG we are able to study origin of many species like protist and find out the biogeographic distribution of various microbes. They also help in detection of new viral strains and species and through understanding cell heterogeneity, viral replication and infection can be fully understood.
Conclusion
As a whole SCG has revealed to be a very powerful tool in the field of life sciences. It has proved to be valuable in almost every field of biology. It is useful in diagnostic, building phyletic lineages, profiling rare cells and especially in cancer biology. It has allowed us to understand various cells and cell types and associate them with specific functions. Although, a lot of work needs to be done in SCG, but still this field has a lot of potential. Its many possible applications include understanding of cell plasticity, cell differentiation patterns on tumor cells and understanding prokaryotic diversity. But more research is needed to understand complex and dynamic population. In addition to this, computational methods are essential and they should be developed for data analysis.

