BioPython
BioPython is a package of freely available tools for biological computation written in the Python programming language. BioPython has various modules that allow you to perform different tasks on your biological data. Hence by using the modules available in the BioPython package you won’t need to write long lines of codes to perform a specific task on your biological data instead you can just call the built-in functions available in BioPython and perform those tasks.
BioPython Installation
In order to install the BioPython package you need to perform the following steps:
Step 1: Open CMD (command prompt) on windows or terminal in the case of macOS or Linux operating system.
Step 2: Write the command: pip install biopython, and press enter in order to call in the pip function.
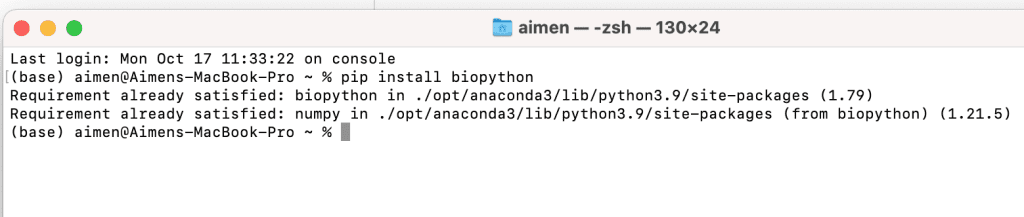
Step 3: To verify if the BioPython package is successfully installed, open the IDLE program which is a built-in interpreter in the Python programming language.
Step 4: Enter the command: from bio.seq import seq, and run it.
Step 5: If it shows no error after running the command that means BioPython has been successfully installed and it is ready to work.
Step 6: Open Visual Code or Jupyter Notebook and write the command: bio.seq import seq, and execute it.
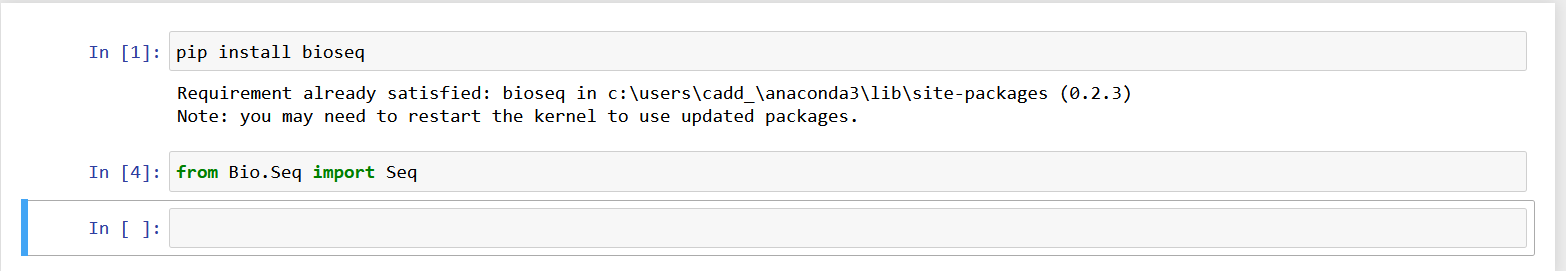
Step 7: If it displays no error on execution that means that your module is ready and you can start working on it.
This way the BioPython package will be successfully installed on your system.