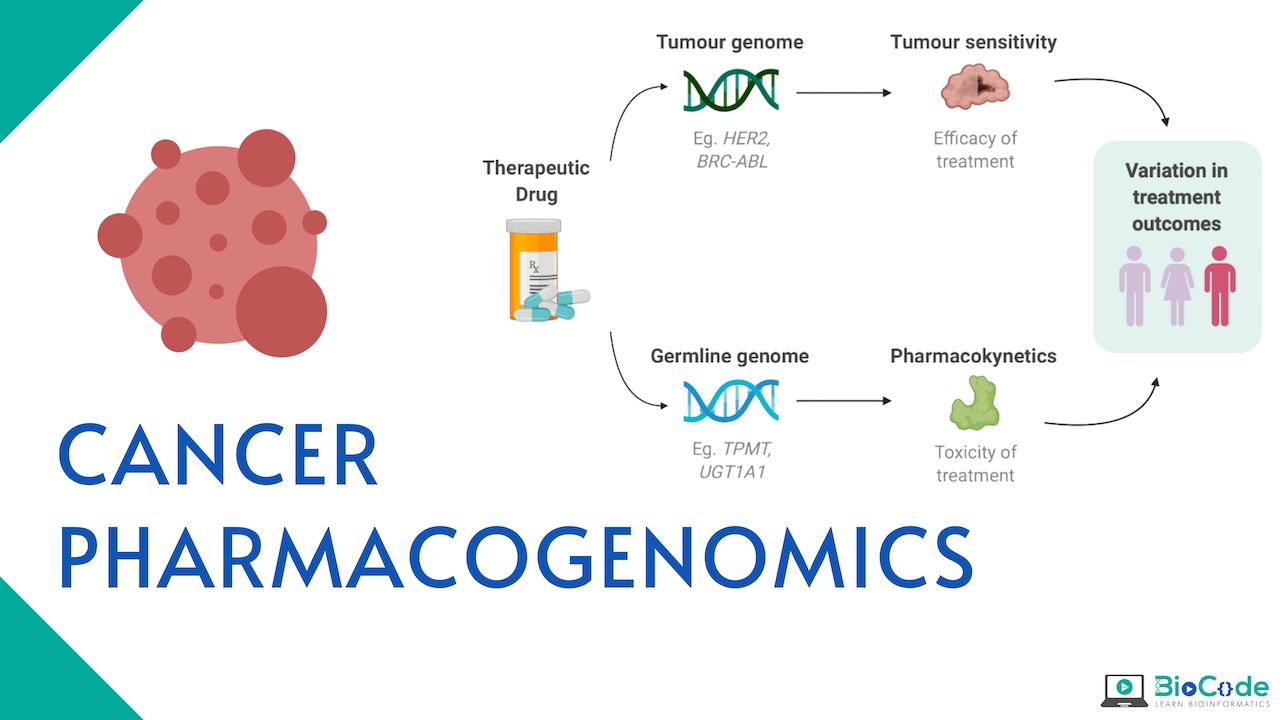
What is Cancer Pharmacogenomics?
Pharmacogenomics is the study of genetic factors that influence drug efficacy and toxicity. These studies aim to elucidate the genetic variation that is present across individuals affecting pharmacodynamics and pharmacokinetics. If the association is found to be reproducible with a large effect size between genotype and drug-induced phenotype, then this knowledge can be used for the benefit of the patient. This is quite important in oncology as cancer is the principal cause of morbidity and death in many industrialized nations and inappropriate treatment can have a life-threatening impact, increasing the number of deaths. Cancer Pharmacogenomics is the precise personal therapy for cancer patients which is recommended based on the patient’s response to the drug.
Terms to understand
Efficacy
In oncology, it refers to tumor response, progression-free survival, and overall survival of an individual.
Pharmacokinetics
It is the effect induced on a drug by the body i.e., the mechanism through which drugs are absorbed, distributed, metabolized, and eliminated by the body.
Pharmacodynamics
The effect of the drug on the body: that is, drug targets and mechanisms of action.
Targeting genomics and transcriptomics in Cancer Pharmacogenomics
A new era of Cancer pharmacogenomics began with the development of transcriptome and editing technologies. These technologies have enables us to identify cancer alterations with high-throughput sequencing technologies. Integration of these techniques has revolutionized biomedical and cancer research. Large-scale international projects like Pan-Cancer Analysis of Whole Genomes and The Cancer Genome Atlas are accurately identifying actionable mutations and extensive tumor heterogeneity. These finding has enabled new drug development strategies. In addition, novel drug discovery can also be done through targeted genome manipulation through the CRISPR system by using dual RNA-guided DNA endonuclease enzyme Cas9 (CRISPR-Cas9). The research has revealed that remarkable, genetic, genomic, and transcriptional heterogeneity exists in individuals. This finding has raised the need to develop predictive biomarkers and novel drugs so that personalized therapy can be optimized.
The fast development in NGS has to allow the incorporation of targeted NGS (tNGS) in both fundamental and translational research through:
Basket trials and clinical implications
Whole-exome sequencing (WES)
Whole-genome sequencing (WGS)
RNA-sequencing (RNA-seq)
The basketball trials are based on the same druggable mutation that is present across different types of cancer and also available approved targeted drugs that match specific onco-targets. On the other hand, WGS and WES with or without RNA-seq have led to the discovery of novel cancer genes and among them, few are identified as new onco-targets for many types of cancer. For instance, a large pan-cancer study was carried out with about 9500 tumor patients from the Cancer Genome Atlas. They have identified a total of 299 cancer driver genes among which about 59 are novel genes. After a comprehensive analysis of databases, considering FDA-approved therapies, clinical trials, and also taking into account the published clinical evidence, it was shown that about 57% of the group has a potentially druggable mutation. Similarly, another reliable publication on WGS analysis of 183 melanoma patients has discovered a significant number of mutated genes including SF3B1, CDKN2A, BRAF, and NRAS. The most actionable alteration was found in MAPK and phosphoinositol kinase pathway components. Also, an important revelation was the fact that many genes were affected by recurrent mutation in non-coding regions, thus showing that they have a strong functional impact like the coding mutations. Furthermore, this study gives very exciting research data which resulted in the setting of many clinical trials to evaluate the potential for new drug discovery.
Another landmark publication of WGS in 2500 patients of Pan-Cancer Analysis of Whole Genomes collaboration provided, for the first time, precise identification of comprehensive driver structural variance and point mutations that were driving personalized tumorigenesis. The most recurrent alterations were:
Coding Point mutations (76%)
Copy number mutations (73%)
Genomic rearrangements (26%)
Non-coding mutations (25%)
On average, almost 4 t mutations were present in the tumor to drive tumorigenesis. The researchers also elucidated that a large number of individual tumors have a specific signature of many mutation types and almost 50% of cancer genes have different combinations of driver mutation classes. Thus, this study for the first time bridged the gap between research and clinical precision medicine by providing an evidence-based strategy to develop a personalized drug. Also, remarkably, in most of the patients, actionable mutations were found to be 80%. Generally, actionable mutations were present in 625 tumors out of which point mutations were 331 while structural copy number mutations were 219 and structural genomic rearrangements were 75. Besides these, approximately 577 biomarkers have been identified that have been targeted by drugs after the approval of the FDA. Also, almost 1678 tumors have been discovered to have druggable events, thus, making them potential therapeutic targets. Finally, in 307 tumors targetable preclinical molecules were found. Thus, this panorama study not only gives information regarding the standard targeting and immunotherapeutic agents but also allows the development of a large drug bank.
In addition to NGS, CRISPR-Cas9 also provides an opportunity to discover onco-targets and the development of target drugs. Currently, Behan and colleagues have used CRISPR-based screening to prioritize drug targets in 30 cancer types cell lines. Many clinical trials are taking place to assess the efficacy of CRISPR.
Somatic Mutations in Cancer pharmacogenomics
Somatic mutations are defined as alterations that are non-hereditary. Mostly, somatic mutations are considered as the driver that describes the cancer subtype or they might be the passengers. This is because tumor samples are not completely pure and they are a combination or mixture of normal and cancer cells. This fact must be taken into account while declaring a somatic mutation in DNA sequence studies.
Tumor samples are mostly small biopsies in which tissue is formalin-fixed and paraffin-embedded (FFPE) and this results in partial degradation of DNA. So, before processing such samples care must be taken to fully ensure that the sample is amenable to genomic analysis. Cancer cells can have heterogeneous mutations i.e., distinct sections of the tumors have different clonal expressions. The current recommendation to deal with heterogeneity is to use ubiquitous mutations present in the trunk of a phylogenetic tree for treatment. Targeted therapies have also been developed for some proteins which are activated through somatic mutations.
To have appropriate targeted therapy for somatic mutations, it is important to understand pathway considerations. For example, in lung cancer the activation of epidermal growth factor receptor (EGFR) signaling takes place through alteration in many different genes present in the pathway in addition to alteration in EGFR itself. The projects like the International Cancer Genome Consortium and the Cancer Genome Atlas are carrying out large genomic studies in thousands of tumors at the level of transcriptome and epigenome in more than 50 cancer types to identify or define somatic driver mutations. In addition to this, these integrative studies of global mRNA and patterns of methylation can unravel novel clinically important diseases subtypes for therapeutic and prognosis management.
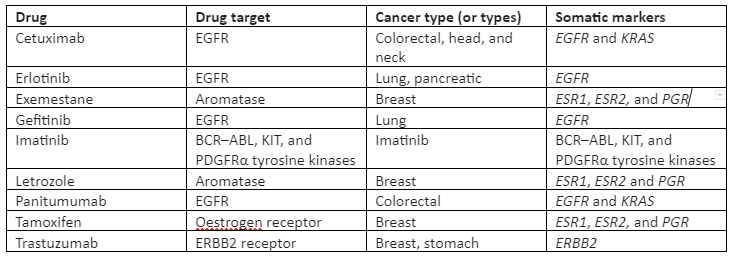
Also, these large-scale projects aim to make the genomic data available to the public, and also this data has already been taken into account for the development of potential inhibitors of genes. In some targeted therapies, treatment efficacy is predicted by somatic mutations. Some of the targeted drugs for somatic mutations are given in Table 1:
Table 1: Somatic mutation targeted FDA-approved drugs
As mentioned, somatic mutations can describe disease subtypes, so they can be crucial covariates in the case where distinct types of tumors are combined in the analysis of germline pharmacogenomics. Also, germline mutations can control that which type of somatic mutation tumor is likely to be acquired. For instance, in a study about squamous cell carcinomas, it was found that the cell carcinomas that arose independently are more alike than the ones present among individuals. This finding demonstrates that germline genetics affects somatic change patterns. Thus, the somatic mutation is used as endophenotypes to test germline genetic variants that are at risk of having a specific type of somatic mutation.
Germline Mutations in Cancer pharmacogenomics
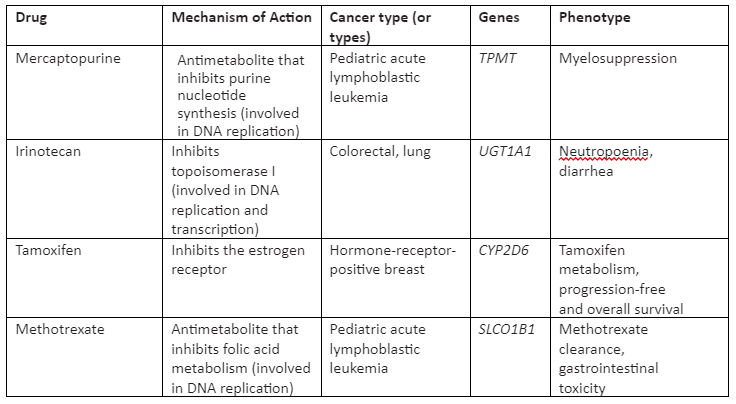
The germline mutations take place in germ cells and are hereditary. The germline mutations that are present in individuals can affect the pharmacodynamics and pharmacokinetics of cancer drugs autonomously, not depending on the disease type. The current treatment which is used is cytotoxic chemotherapy, not used for somatic mutations, that initiate malignant transformation as the driver mutations are not known in many patients. Some of the cytotoxic effects of chemotherapeutic agents are found to be heritable in patients treated with them through studies of cell-line pedigrees. So, variations in toxicity of the drug and its response experienced by cancer patients have made the researchers find out germline mutations that are linked with chemotherapy-induced phenotype. This will enable us to set the dose and toxicity according to the drug response. Some of the cancer-drug for germline mutation with their mechanism of action is given in Table 2:
Table 2: Cancer drugs for germline mutation and associated phenotype
Study Design
In cancer pharmacogenomics, the study design is mostly done with a candidate gene approach. In this approach, variants in known drug targets and drug-metabolizing enzymes are tested for affiliation with the phenotype of interest. Genotyping arrays which contain hundreds of SNPs in identified drug absorption, metabolism, metabolism, and elimination genes (ADME) is very important in candidate gene studies of pharmacogenomics. Besides this, this approach needs prior biological knowledge and can miss the regions which are not known. But this approach has still got merits and advantages in cancer pharmacogenomics in cases where the sample size is small and especially in the presence of pharmacokinetic data. Nevertheless, despite all these advantages and with decreasing cost of sequencing all the efforts should be made for a comprehensive genome-wide analysis.
The ideal structure for pharmacogenomics is provided by the clinical trials due to their constant dosing of drugs and collection of phenotypes. The phases of trials are:
Phase 1 trials: Determines maximum tolerable dose of new drug
Phase 2 trial: Measure effectiveness of the drug.
Phase 3: Genome-wide association studies (GWAS)
The sample size for phase 1 and phase 2 are small, mostly less than 100 individuals, and thus they are not suitable for genome-wide pharmacogenomics discovery studies, but they are useful in candidate gene studies. The sample size of phase three is large as it consists of hundreds or thousands of patients and is hence useful for GWAS. Also, cancer pharmacogenomics studies having a perspective can be designed distinctly from clinical trials but they must be taken into account for dosage, phenotype effects and covariate collection procedure.
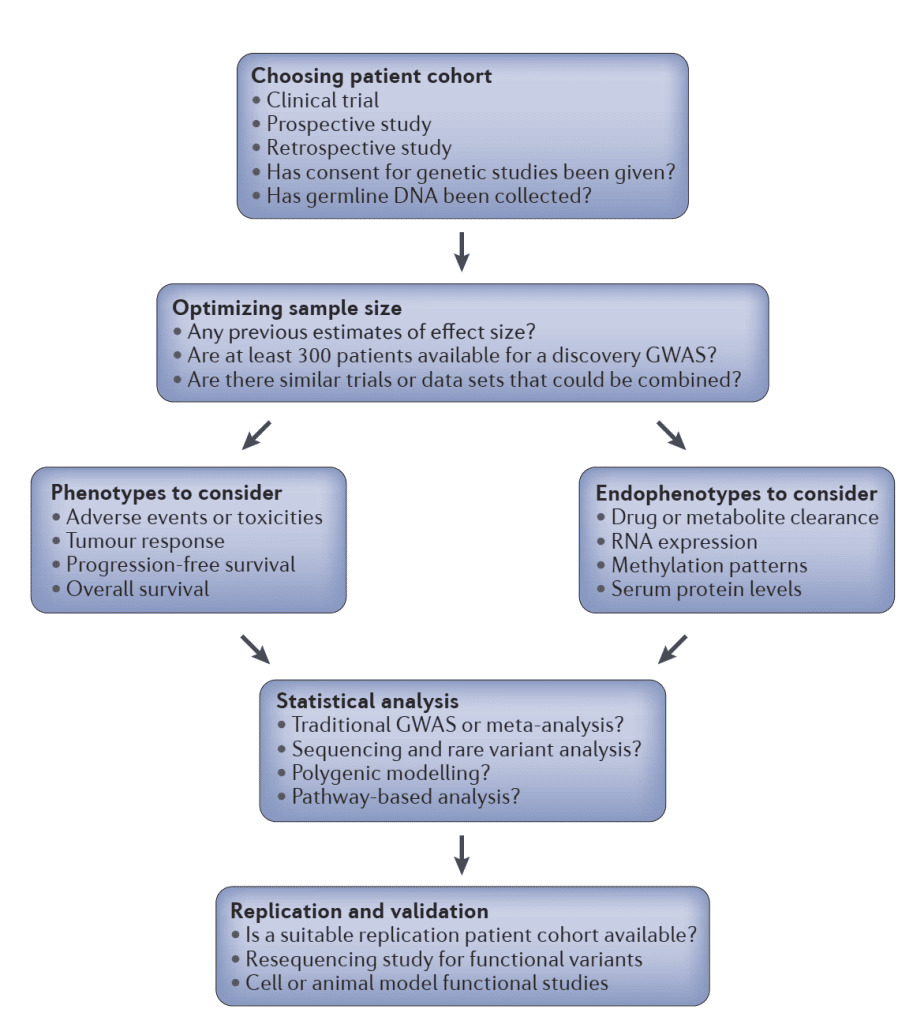
Overview of cancer pharmacogenomics study design
Challenges.
The future of cancer pharmacogenomics depends upon the accurate characterization of genome and transcriptome-wide cancer mutations. This allows us to understand the role of coding and noncoding variants in the regulation of gene expression and drug response through deregulation of cis and trans-regulatory networks. Effective target drug development needs precise clinical trials through techniques like NGS and CRISPR Cas9. These next-generation techniques not only allow us to identify biomarkers and potential targeted drug sites but also take part in the development of innovative agents that can target key regulators in transcriptional pathways. But still, work needs to be done and more research is required in this domain.