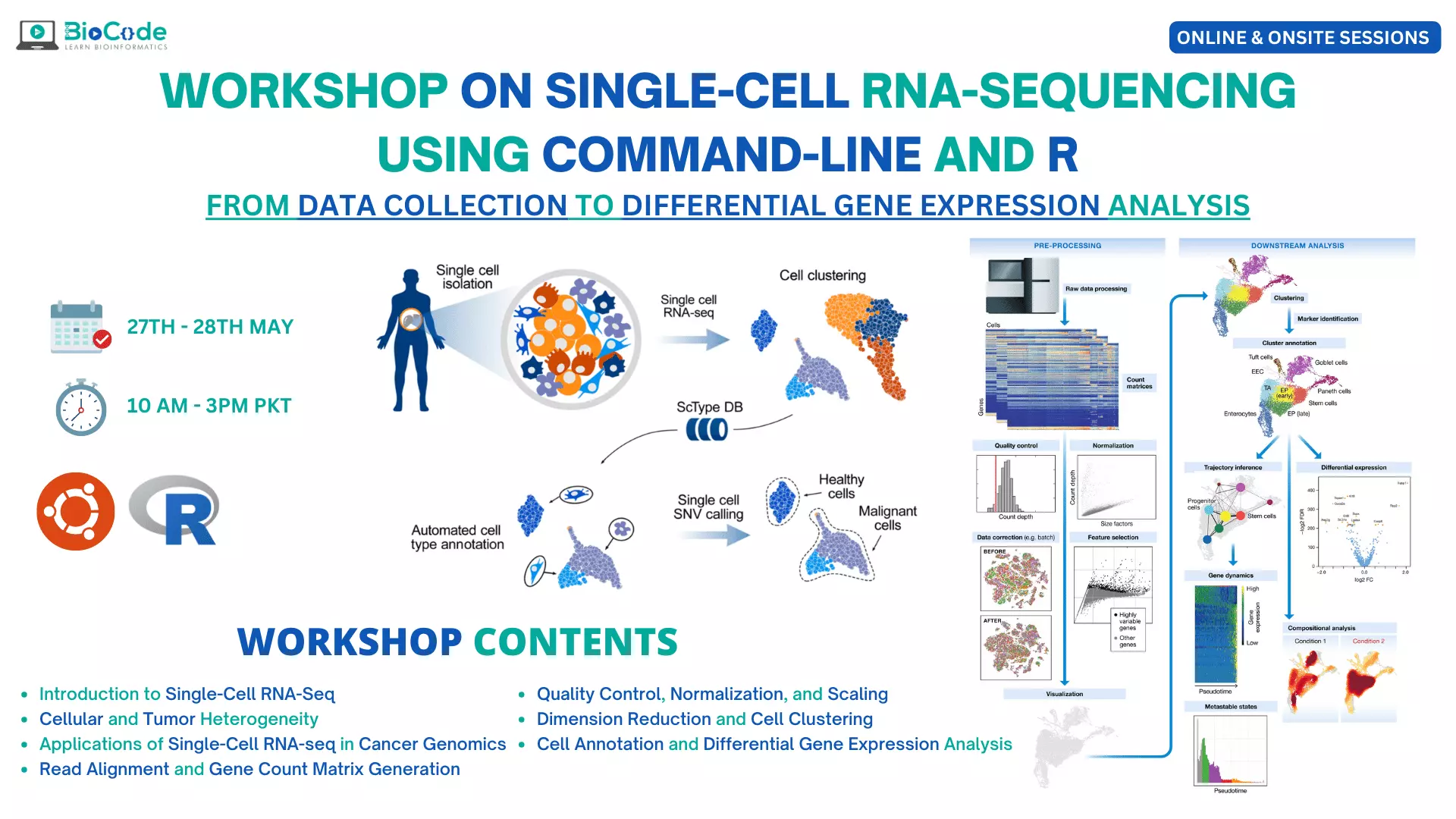
Single-cell RNA sequencing (scRNA-seq) is a technology that allows researchers to study the gene expression patterns of individual cells in a population. Traditional bulk RNA-seq measures the average gene expression across all cells in a sample, which can mask important biological heterogeneity between different cell types or individual cells within a population. scRNA-seq, on the other hand, enables the measurement of gene expression in individual cells, allowing researchers to identify rare or previously unknown cell types and study the molecular processes that underlie cellular heterogeneity.
scRNA-seq has revolutionized the field of bioinformatics by allowing researchers to study the characteristics of individual cells in detail. scRNA-seq can be used to study gene regulation and epigenetics, by analyzing the expression of individual genes in different cells and under different conditions. scRNA-seq has also been used to study disease mechanisms and identify biomarkers for various diseases, including cancer, neurological disorders, and autoimmune diseases. scRNA-seq has been used in drug discovery thereby helping researchers to develop more personalized therapies.
Therefore, BioCode is offering a hands-on workshop on Single-Cell RNA-Sequencing using Command-Line and R in which the participants will learn how to analyze single-cell RNA-sequencing data using the command-line interface and R programming language. Throughout the workshop, participants will be given hands-on exercises to practice what they will learn. The workshop is divided into several sessions, each focusing on a particular aspect of the analysis process.
The workshop is intended for researchers, students, and bioinformatics professionals who are interested in scRNA-seq data analysis and want to learn how to use command-line tools and R to analyze scRNA-seq data. Throughout the workshop, participants will have the opportunity to work with and analyze a sample scRNA-seq dataset. The workshop will also cover best practices for scRNA-seq analysis, including how to deal with technical noise, batch effects, and other common issues that arise when analyzing scRNA-seq data.
By the end of the workshop, participants will have gained a solid understanding of scRNA-seq analysis and the importance of scRNA-seq in biological research. They will have the practical skills and knowledge necessary to analyze scRNA-seq data, and participants will also be able to apply these techniques to their own research questions.
Session 1: Introduction to Single-Cell RNA-Seq
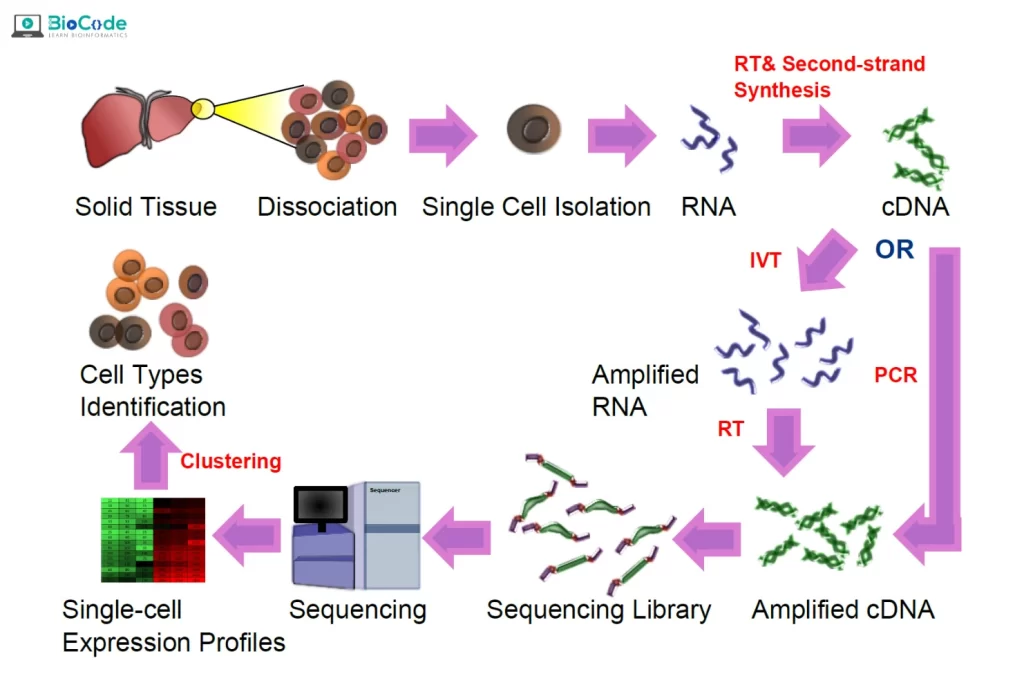
Description: In this session, participants will learn about the basics of scRNA-seq technology. They will also be introduced to the key concepts and terminology used in scRNA-seq analysis. Participants will learn about the importance of scRNA-seq in studying cellular and tumor heterogeneity.
Learning Outcomes: Upon completion of this section, participants will be able to:
- Explain the Basics of ScRNA-Seq.
- Describe the ScRNA-Seq Pipeline.
- Discuss the Importance of ScRNA-Seq
Session 2: Cellular and Tumor Heterogeneity
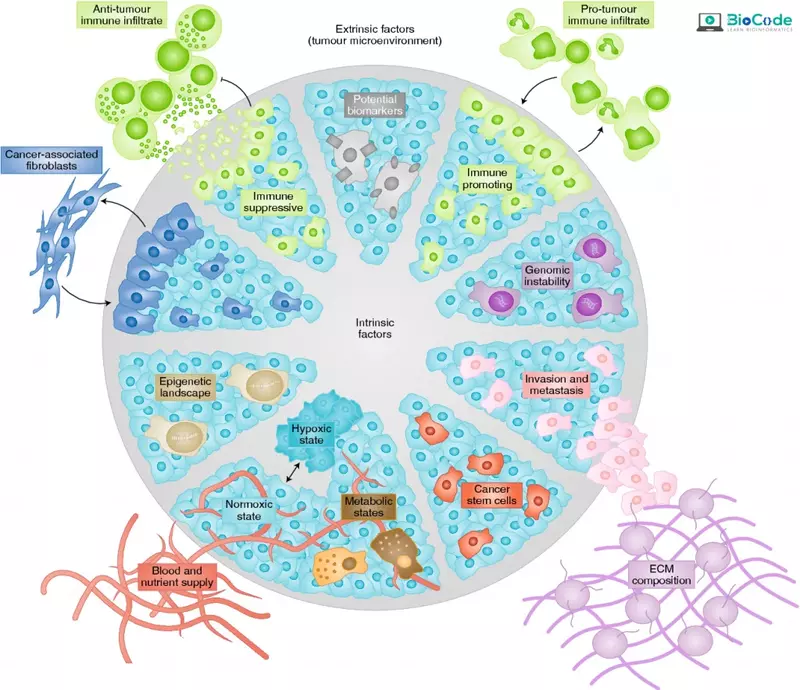
Description: In this session, participants will explore the role of cellular and tumor heterogeneity in scRNA-seq analysis. They will learn about the different types of cells and cell populations that can be identified using scRNA-seq, and the importance of understanding cellular heterogeneity in biological research.
Learning Outcomes: Upon completion of this section, participants will be able to:
- Explain Cellular and Tumor Heterogeneity.
- Describe the Cell Types and Cell Populations Which Can Be Identified Using ScRNA-Seq.
- Discuss the Importance of Cellular Tumor Heterogeneity.
Session 3: Applications of Single-Cell RNA-seq in Cancer Genomics
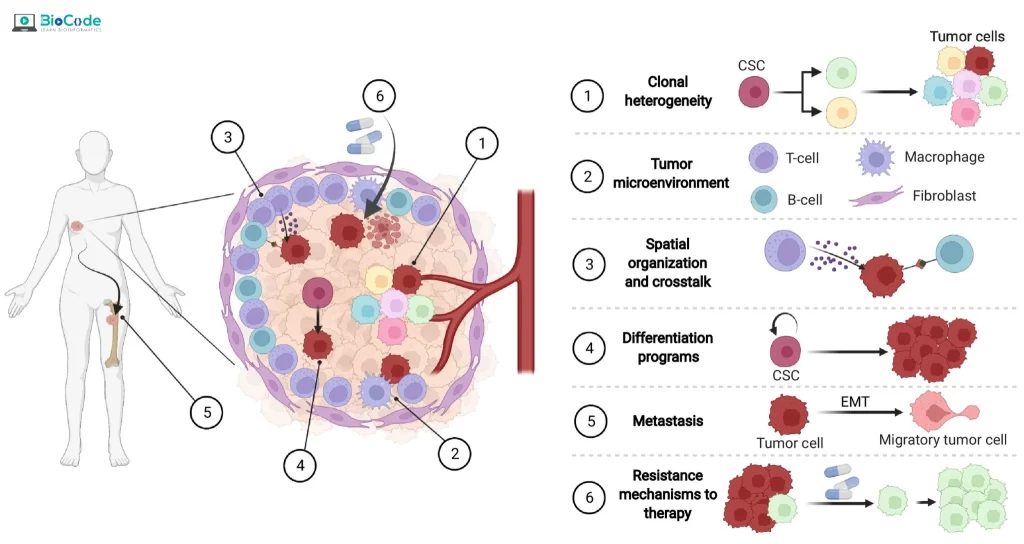
Description: In this session, participants will focus on the applications of scRNA-seq in cancer genomics research. Participants will learn how the identification of single cells helps in the diagnosis and prognosis of various cancers. They will also learn about the different research questions that can be addressed using scRNA-seq data.
Learning Outcomes: Upon completion of this section, participants will be able to:
- Discuss the Applications of scRNA-seq in Cancer Genomics.
- Describe the Cancer Cell Types.
- Study Cancer Cells Using scRNA-Seq.
Session 4: Read Alignment and Gene Count Matrix Generation
Description: In this session, participants will learn the technical aspects of scRNA-seq analysis, including read alignment and gene count matrix generation. Participants will learn how to align scRNA-seq reads to a reference genome and how to generate a gene count matrix from the aligned reads.
Learning Outcomes: Upon completion of this section, participants will be able to:
- Perform Read Alignment.
- Generate a Gene Count Matrix.
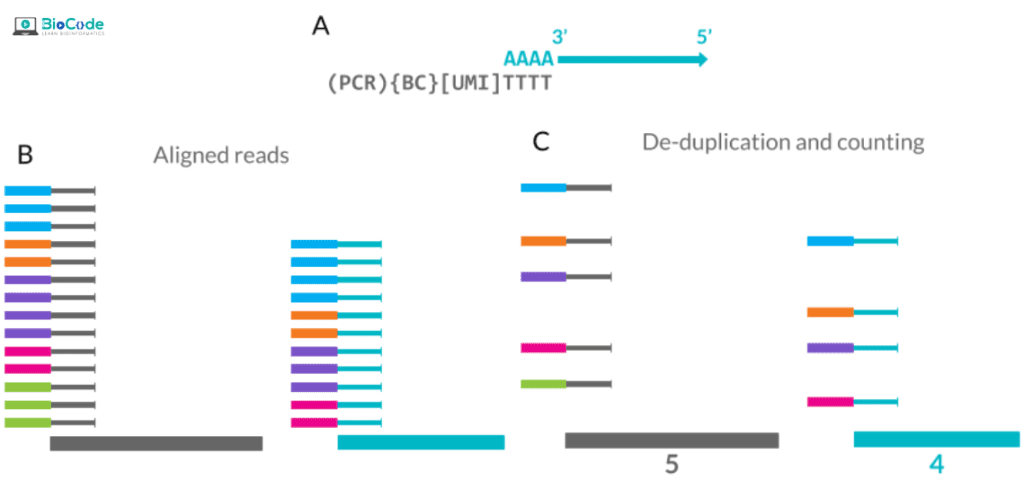
Session 5: Quality Control, Normalization, and Scaling
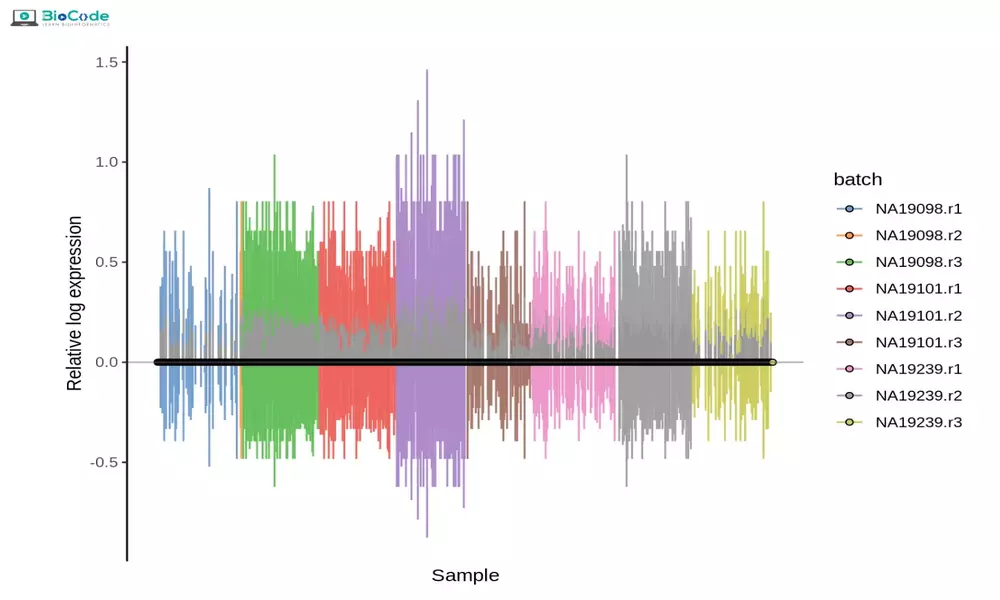
Description: In this session, participants will learn how to perform quality control on scRNA-seq data, normalize it and apply scaling techniques in order to remove technical variations. They will learn how to identify potential outliers and technical issues, as well as how to reduce technical noise and improve the accuracy of downstream analyses.
Learning Outcomes: Upon completion of this section, participants will be able to:
- Perform Quality Control.
- Perform Normalization & Scaling.
Session 6: Dimension Reduction and Cell Clustering
Description: In this session, participants will learn about dimension reduction and cell clustering. They will explore various dimensionality reduction techniques such as UMAP, PCA and t-SNE and learn how to use them to visualize and explore their scRNA-seq data. Participants will also learn how to perform clustering analysis on scRNA-seq data to identify cell populations.
Learning Outcomes: Upon completion of this section, participants will be able to:
- Explain Dimension Reduction and Cell Clustering.
- Perform Dimension Reduction.
- Identify Cell Populations Through Cell Clustering Analysis.
- Visualize and Explore scRNA-Seq Data.
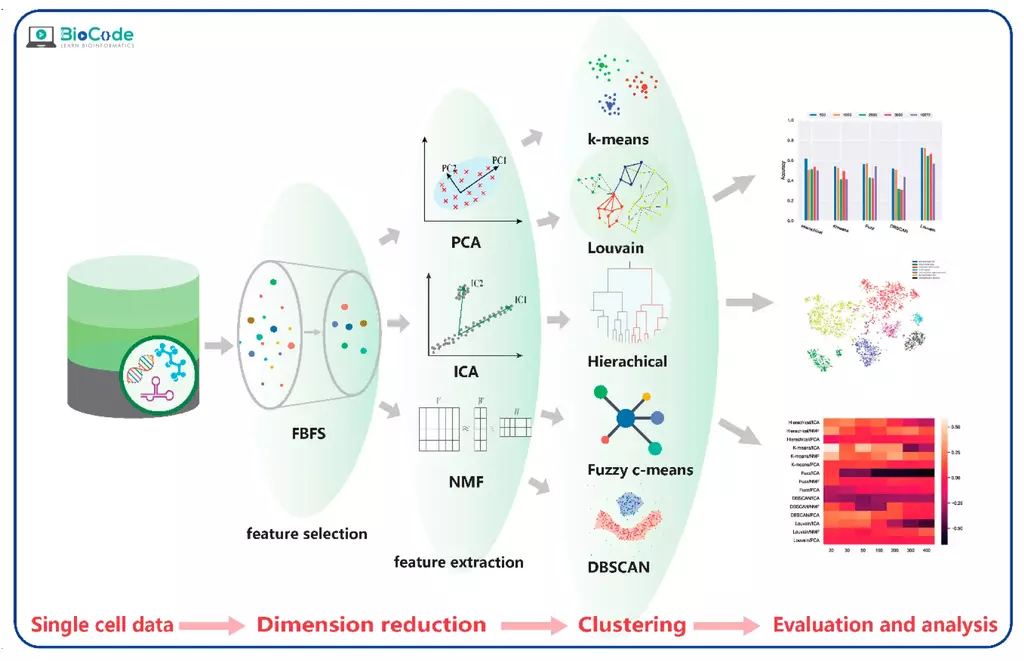
Session 7: Cell Annotation and Differential Gene Expression Analysis

Description: In this session, participants will learn about cell annotation and differential gene expression analysis. Participants will learn how to identify cell types and annotate their scRNA-seq data using gene expression signatures. They will also learn how to perform differential gene expression analysis to identify genes that are differentially expressed between different cell populations.
Learning Outcomes: Upon completion of this section, participants will be able to:
- Explain Cell Annotation and Gene Expression Analysis.
- Identify Cell Types in scRNA-Seq Data.
- Annotate ScRNA-Seq Data Using Gene Expression Signatures.
- Perform Differential Gene Expression Analysis.
Date: 27-28th May, 2023
10 AM PKT to 3 AM PKT (if you can’t join, you will receive recorded sessions)
Recorded sessions will be provided, you can join the course without worrying about the time.
Session is available both online and on-site.
Price: 100.00 USD
