Hands-on: NGS (Whole Exome Sequencing) Variant Calling Using Python
About Course
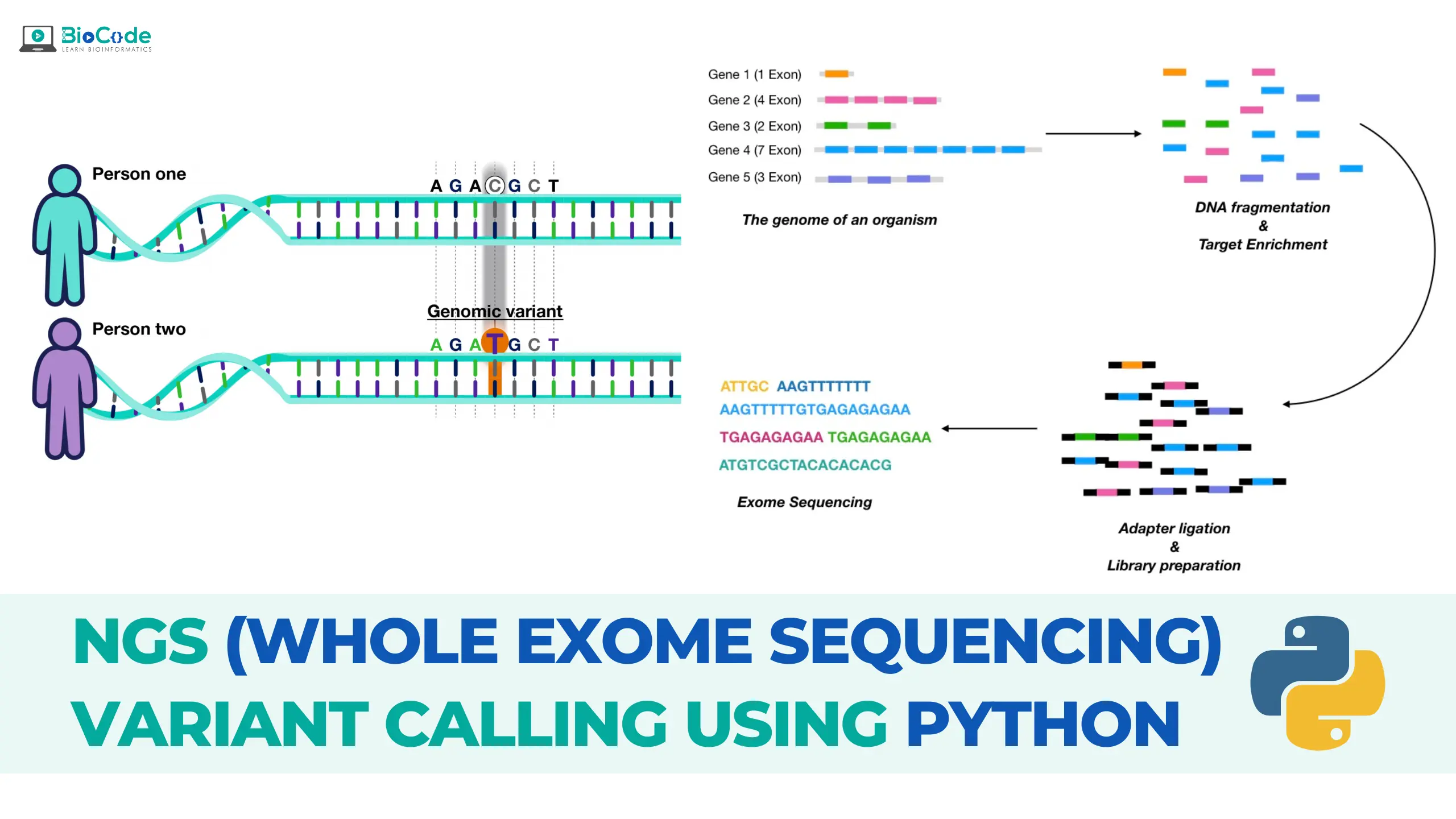
Hands-on: NGS (Whole Exome Sequencing) Variant Calling Using Python
The primary goal of this course is to thoroughly equip students with the necessary skills and knowledge in Next Generation Sequencing (NGS) and whole exome data analysis. It covers the foundational aspects of NGS while also providing hands-on training in variant calling applicable to any genome or organism. Throughout the course, students will learn to identify and analyze variations, including single nucleotide polymorphisms (SNPs) and insertions/deletions (indels), within raw datasets.
The course commences by delving into the fundamentals of NGS, elucidating various platforms, sequencing technologies, and their respective advantages and limitations. Additionally, students will acquire proficiency in obtaining raw whole exome datasets tailored to any organism or disease of interest.
Once students have acquired the requisite data, the course proceeds with practical training encompassing quality control, trimming, mapping against a reference genome, post-alignment quality assessment, read recalibration, and variant identification within disease/patient samples to generate Variant Call Format (VCF) files. Furthermore, students will be introduced to a myriad of tools aimed at filtering and annotating VCF files, thereby ensuring the accuracy and reliability of their analytical outcomes.
Throughout the course duration, students will engage extensively with authentic NGS data, thereby honing their abilities to troubleshoot common challenges and optimize analysis pipelines to accommodate diverse data types. By the culmination of the course, participants will emerge with a proficient understanding of NGS and whole exome data analysis, thereby empowering them to undertake advanced research endeavors or industry projects within the realm of genomics.
The course is structured into three distinct categories:
- Introduction to NGS and Whole-Exome Sequencing:
- This segment provides a comprehensive introduction to whole exome sequencing (WES), elucidating its foundational principles and its applicability to comprehensive genomic analysis. Topics covered include distinctions between haploid and diploid organisms in variant calling, the significance of ploidy in disease research, and an overview of germline versus somatic mutations. Additionally, participants will be acquainted with the nuances of single nucleotide polymorphisms (SNPs), large-scale genome variations, and the pivotal role of Copy Number Variations (CNVs) in the manifestation of various diseases.
- Hands-on Whole-Exome Variant Calling:
- In this category, participants will undergo intensive training in employing the Genome Analysis Toolkit (GATK) pipeline for variant calling. Practical sessions will entail hands-on experience in retrieving raw whole exome sequencing (WES) data, assessing read quality, conducting mapping procedures against a reference genome, executing variant calling utilizing Freebayes, and performing subsequent variant filtration. Moreover, participants will gain proficiency in utilizing tools such as VCFtools and SnpSift for variant file manipulation and quality filtering, thereby enriching their analytical capabilities.
- Additional Downstream Analysis:
- The final category focuses on downstream functional enrichment analysis techniques, aimed at unraveling the biological significance of identified variants. Participants will engage in hands-on practice sessions, wherein they will undertake gene ontology analysis utilizing topGO and enrichR, alongside pathway analysis employing renowned resources such as KEGG, PANTHER, and Reactome.
Upon successful completion of the course, participants will emerge equipped with a comprehensive understanding of NGS and whole exome sequencing (WES), adeptness in variant calling methodologies, and proficiency in employing essential bioinformatics tools for robust data analysis.
Course Content
In-depth Introduction to NGS and Whole-Exome Sequencing
-
In-depth Introduction to NGS and Variant Calling
20:12 -
Fundamentals of Whole Exome Sequencing: A Comprehensive Sequencing Approach
26:16 -
Haploid vs. Diploid Organisms for Variant Calling
07:26 -
Ploidy and Its Significance in Disease Research
14:58 -
A Bioinformatics Approach Towards Germline vs. Somatic Mutations
14:30 -
Single Nucleotide Polymorphisms: An Introduction to SNPs and Their Types
31:22 -
Structural Variations: Large Scale Variations in Genomes
08:55 -
Copy Number Variations: Role of CNVs in Diseases
10:29
Hands-on Whole-Exome Variant Calling Using Python-Based Scripts
Additional Downstream Analysis
Additional Lectures
Earn a certificate
Add this certificate to your resume to demonstrate your skills & increase your chances of getting noticed.

Student Ratings & Reviews

