Hands-on: NGS (Whole Exome Sequencing) Variant Calling Using Linux
About Course
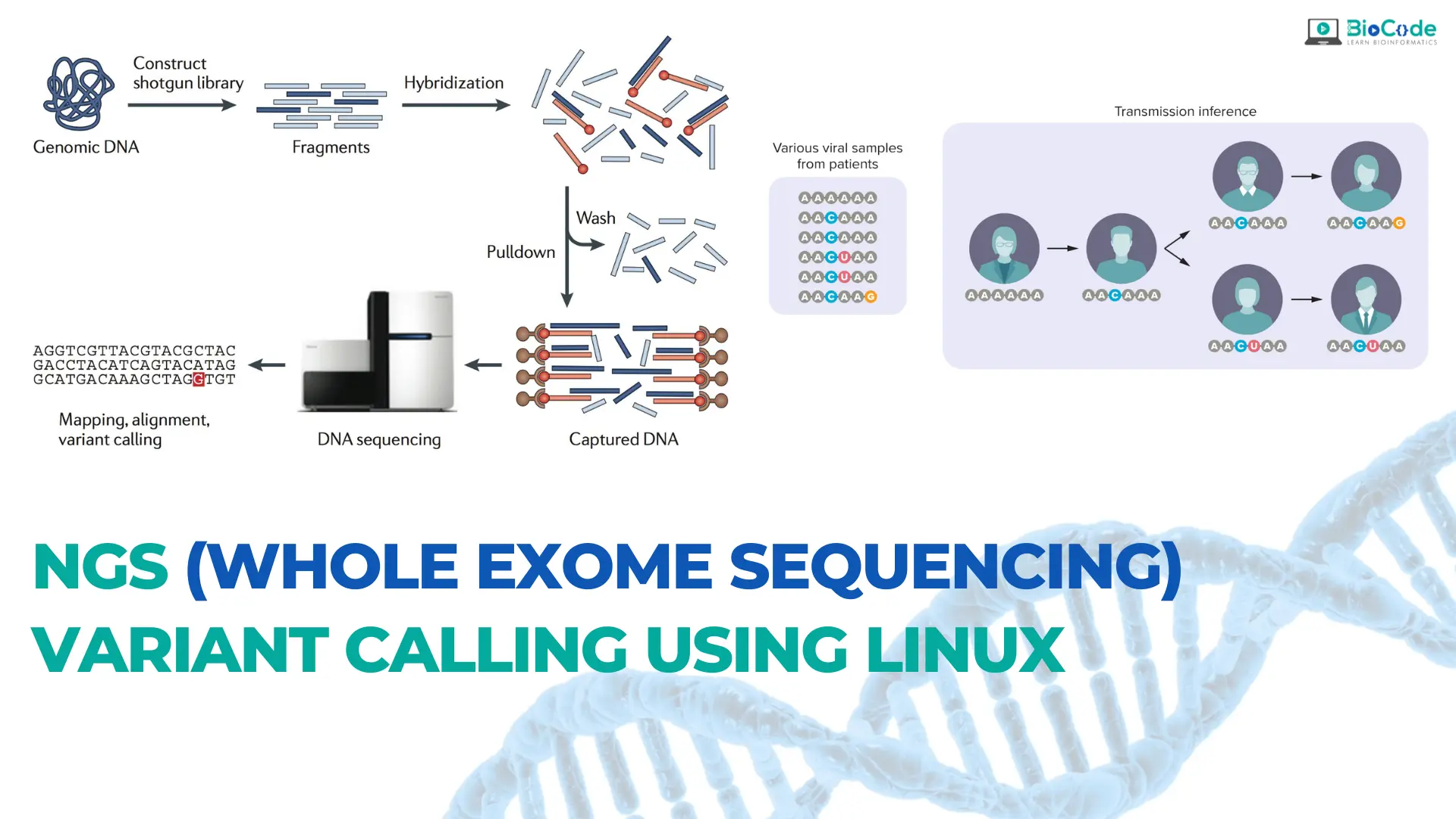
This comprehensive course in NGS/whole exome data analysis is designed to provide students with a thorough understanding of the basics of NGS and hands-on training in whole-genome variant calling on any genome or organism. Throughout the course, students will learn the complete process of identifying and analyzing variations, including substitutions (SNPs) and insertions/deletions (indels) in a raw dataset.
The course will start with the basics of NGS, including how it works, the different platforms and sequencing technologies available, and the advantages and limitations of NGS. Students will also learn how to obtain raw whole exome datasets for any organism or disease.
Once the students have obtained the data, the course will provide practical training on how to do quality control, trimming, mapping against a reference genome, post-alignment quality control, recalibration of the reads, and finally, how to identify variants in the disease/patient samples to obtain VCF files. Students will learn how to use various tools to filter and annotate these VCF files, ensuring the accuracy and reliability of their results.
Throughout the course, students will gain hands-on experience working with real NGS data and learn how to troubleshoot common issues and optimize their analysis pipeline for different types of data. By the end of the course, students will have a thorough understanding of NGS and whole exome data analysis and will be well-equipped to apply their skills to research or industry projects in the field of genomics.
This course offers a comprehensive introduction to Next Generation Sequencing (NGS) and Whole-Exome Sequencing (WES) with a focus on variant calling. The course is divided into three categories.
Category 1: In-depth Introduction to NGS and Whole-Exome Sequencing
This category provides an in-depth introduction to NGS and WES. Participants will learn about the fundamentals of WES and the comprehensive sequencing approach it offers. They will also learn about haploid vs. diploid organisms for variant calling, ploidy and its significance in disease research, and a bioinformatics approach towards germline vs. somatic mutations. The category also covers an introduction to Single Nucleotide Polymorphisms (SNPs) and their types, as well as large-scale Hands-on: NGS (Whole Exome Sequencing) Variant Calling Using Linux 3 variations in genomes and the role of Copy Number Variations (CNVs) in diseases.
Category 2: Hands-on Whole-Exome Variant Calling
In this category, participants will learn how to use the Genome Analysis Toolkit (GATK) pipeline for variant calling. They will gain hands-on experience retrieving raw WES data, understanding and filtering low-quality reads, and mapping reads against a reference genome. The category also covers utilizing Freebayes for variant calling and variant filtration, as well as using VCFtools for variant file formats. Participants will learn how to filter out lowquality variants using SnpSift, and how to annotate SNPs with biological knowledge using SnpEff for prediction of variant effects.
Category 3: Additional Downstream Analysis
In this category, participants will learn about downstream functional enrichment analysis. They will gain hands-on experience with gene ontology analysis using topGO and enrichR, as well as pathway analysis using KEGG, PANTHER, and Reactome.
By the end of this course, participants will have a comprehensive understanding of NGS and WES, and the ability to perform variant calling and downstream analysis. They will also gain experience with bioinformatics tools and approaches that are essential for NGS and WES data analysis.
Course Content
In-depth Introduction to NGS and Whole-Exome Sequencing
-
In-depth Introduction to NGS and Variant Calling
20:12 -
Fundamentals of Whole Exome Sequencing: A Comprehensive Sequencing Approach
26:16 -
Haploid vs. Diploid Organisms for Variant Calling
07:26 -
Ploidy and Its Significance in Disease Research
14:58 -
A Bioinformatics Approach Towards Germline vs. Somatic Mutations
14:30 -
Single Nucleotide Polymorphisms: An Introduction to SNPs and Their Types
31:22 -
Structural Variations: Large Scale Variations in Genomes
08:55 -
Copy Number Variations: Role of CNVs in Diseases
10:29
Hands-on Whole-Exome Variant Calling
Additional Downstream Analysis
Additional Lectures
Earn a certificate
Add this certificate to your resume to demonstrate your skills & increase your chances of getting noticed.

Student Ratings & Reviews

