End-to-End RNA-Seq Data Analysis With Python-Based Pipeline
About Course
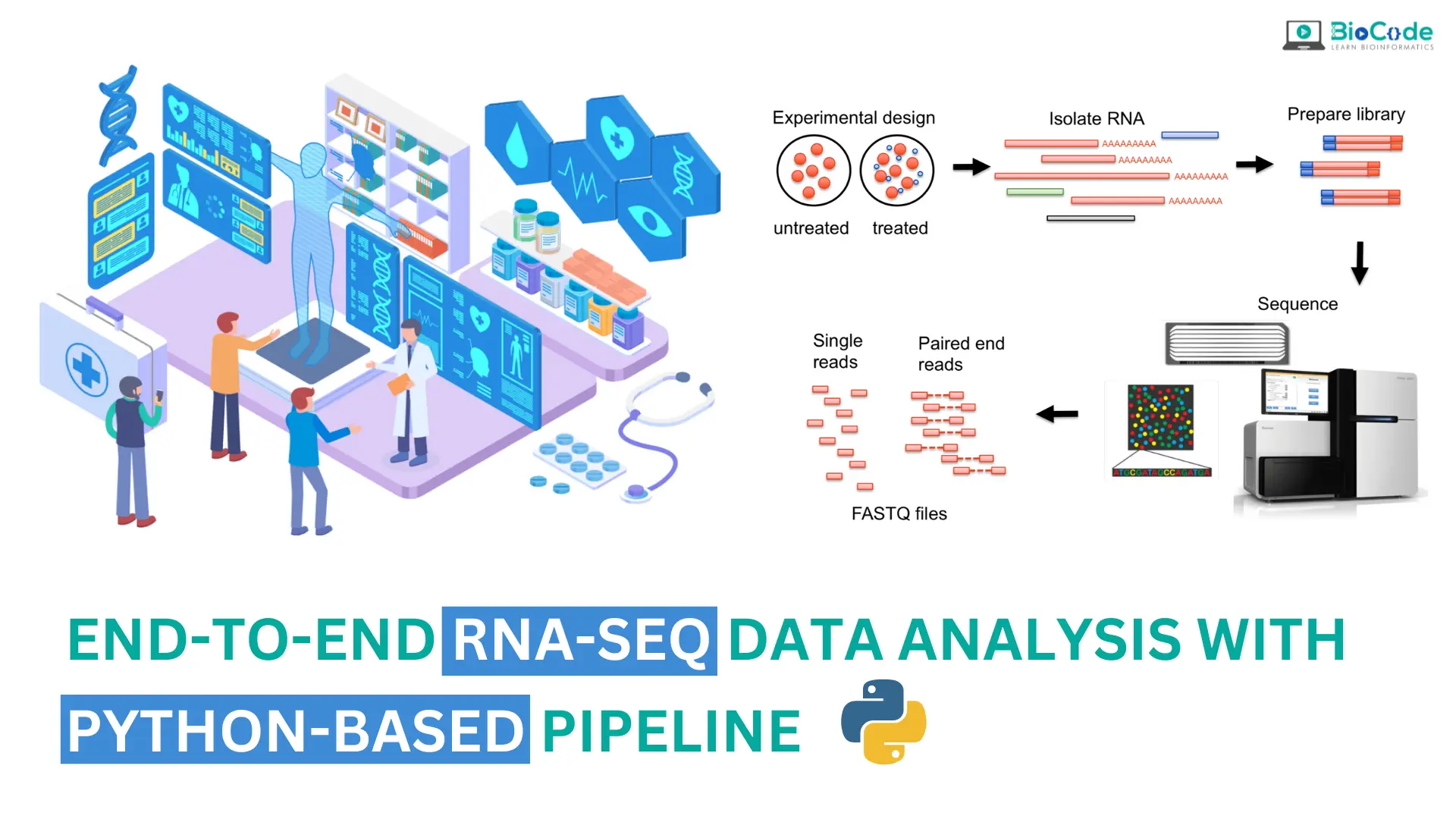
“End-to-End RNA-Seq Data Analysis With Python-Based Pipeline” is a comprehensive course designed to equip learners with the skills and knowledge necessary to conduct thorough RNA-Seq data analysis using a Python-based pipeline. From conceptual understanding to practical implementation, this course covers all facets of RNA-Seq data analysis, making it ideal for students, researchers, and professionals in bioinformatics, genomics, and computational biology.
The course begins with an exploration of fundamental concepts in RNA-Seq data analysis, including sequencing technology, quality control measures, and normalization techniques. Participants will then delve into the preprocessing of raw RNA-Seq data, mastering tasks such as quality assessment, adapter trimming, and read alignment to a reference genome.
Through engaging lectures and hands-on exercises, learners will gain proficiency in differential gene expression analysis using statistical models and data visualization tools. Additionally, participants will become adept in leveraging popular Python libraries for data analysis, including NumPy, Pandas, Matplotlib, and Seaborn.
Real-world RNA-Seq datasets will be utilized throughout the course, providing learners with practical experience and enabling them to apply their newfound skills to a comprehensive case study. Interactive exercises and quizzes will reinforce learning objectives and offer immediate feedback, ensuring a dynamic and effective learning experience.
By the conclusion of the course, participants will have constructed a robust pipeline for RNA-Seq data analysis using Python. They will possess the ability to perform quality control assessments, execute read alignments, conduct differential gene expression analyses, and produce publication-quality visualizations. Equipped with these skills, learners will be well-prepared to undertake RNA-Seq data analyses independently and pursue further studies in bioinformatics, genomics, and computational biology.
No prior programming experience is required to enroll in this course, although a familiarity with the Unix command line and basic statistical concepts is recommended. To get started, participants need only access to a computer with a recent version of Python and the requisite libraries installed.
Enroll today in “End-to-End RNA-Seq Data Analysis With Python-Based Pipeline” and embark on a journey towards mastering RNA-Seq data analysis using Python-based tools!
Course Content
In-Depth Introduction to RNA-Seq
-
44:08
-
Introduction to ArrayExpress
09:56
Genomic Databases for Raw Data
File Formats
RNA-Seq Data Analysis Pipeline (Theoretical & Practical)
Earn a certificate
Add this certificate to your resume to demonstrate your skills & increase your chances of getting noticed.

Student Ratings & Reviews

