Hands-on: Single-Cell RNA-Sequencing Data Analysis Using Command-Line and R [Complete Training]
About Course
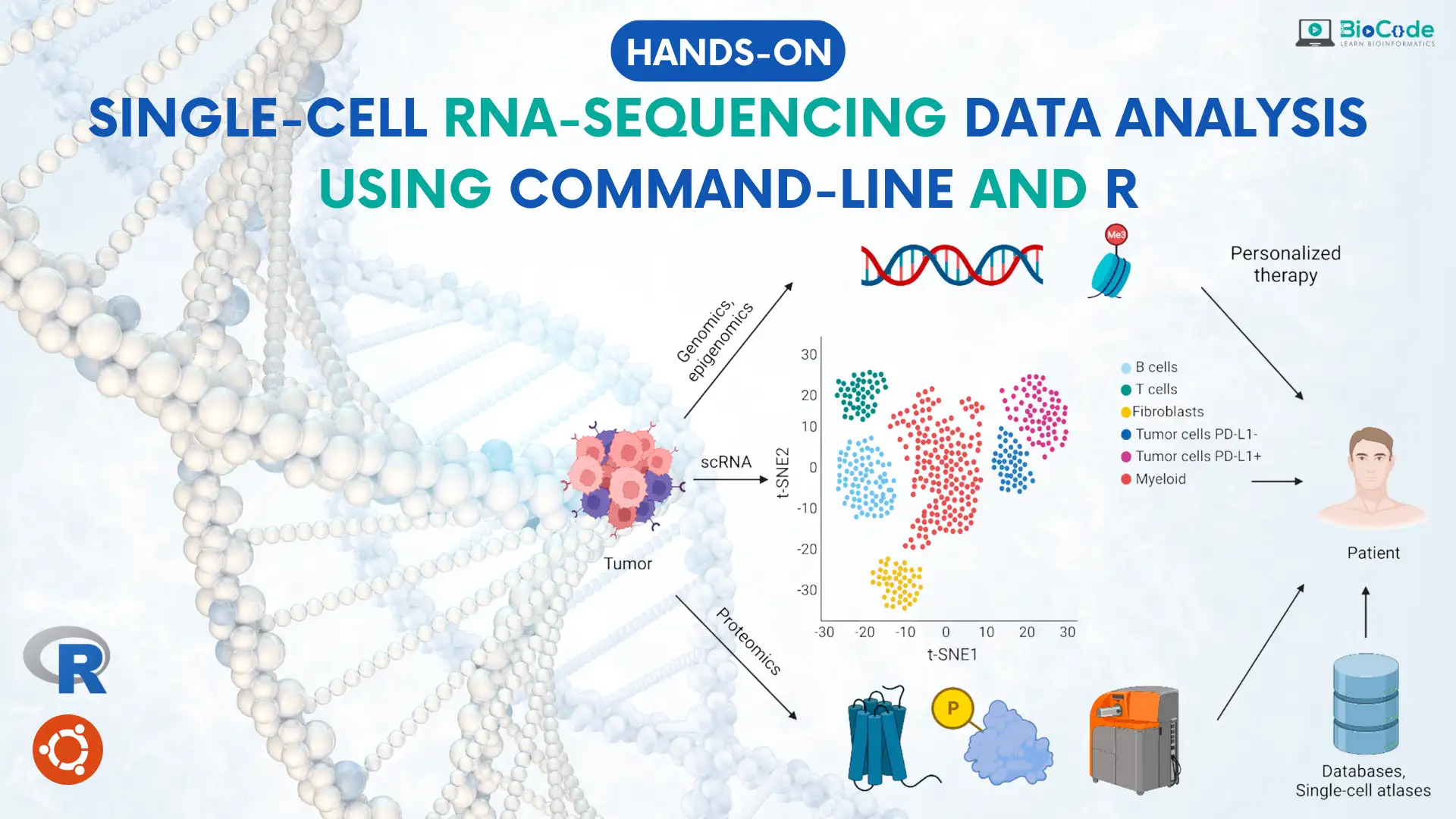
This course provides a comprehensive end-to-end analysis of single cell RNA sequencing (scRNA-seq) data, tailored to guide beginners without prior programming or Linux knowledge. The course emphasizes the use of command-line tools and R packages such as Seurat and scType, and focuses on the fundamental differences between scRNA-seq and bulk RNA-seq, highlighting the necessity for scRNA-seq in investigating cellular and tumor heterogeneity. The course provides a thorough overview of all available scRNA technologies, with particular emphasis on 10x genomics and Smart-seq2, and offers both theoretical and hands-on training for every step in the data analysis pipeline.
Students are instructed on how to process raw scRNA datasets, including UMI, cell barcode filtering, alignment against reference genome, and deduplication and quantification to generate a count matrix. They also learn how to perform quality control on the data, remove unwanted dead cells, normalize and scale the data, perform data imputation, and perform dimension reduction and visualization through UMAP, tSNE, and PCA plots. The course covers cell clustering and subpopulation identification, including cell annotation, and also provides instruction on cell lineage and trajectory analysis. Finally, students learn how to perform differential gene expression analysis, identifying marker genes expressed only in their chosen subpopulations of cells. Overall, this course provides a comprehensive and practical introduction to scRNA-seq analysis for beginners, enabling them to gain the necessary skills to conduct their own analyses.
Course Content
In-depth Introduction to Single Cell RNA-Sequencing, Pipeline and Single-Cell Technologies
-
Introduction to Single-Cell RNA-seq, Its Pipeline and Analysis
23:34 -
Gene Expression and Its Significance
21:42 -
Cellular and Tumor Heterogeneity
09:10 -
Bulk RNA-sequencing vs. Single-Cell RNA-sequencing
21:50 -
Single-Cell RNA-seq Technologies (10x Genomics, Smart-Seq, Drop-seq and more)
23:09 -
Cell Isolation and Cell Lysis Protocols for Single-Cell Genomics
09:53 -
In-depth Introduction to Single-Cell RNA-sequencing Analysis Pipeline
24:45 -
Droplet Technologies: 10x Genomics
14:10 -
Full-Length Transcript Technologies: Smart-Seq2
17:55
Hands-on Single-Cell RNA-seq Data Analysis (From Raw Reads to Cell Subpopulation Identification and DEGs Identification)
Additional Supplementary Lectures
Earn a certificate
Add this certificate to your resume to demonstrate your skills & increase your chances of getting noticed.

