Hands-on: Single-Cell RNA-Sequencing Data Analysis Using Python [Complete Training]
About Course
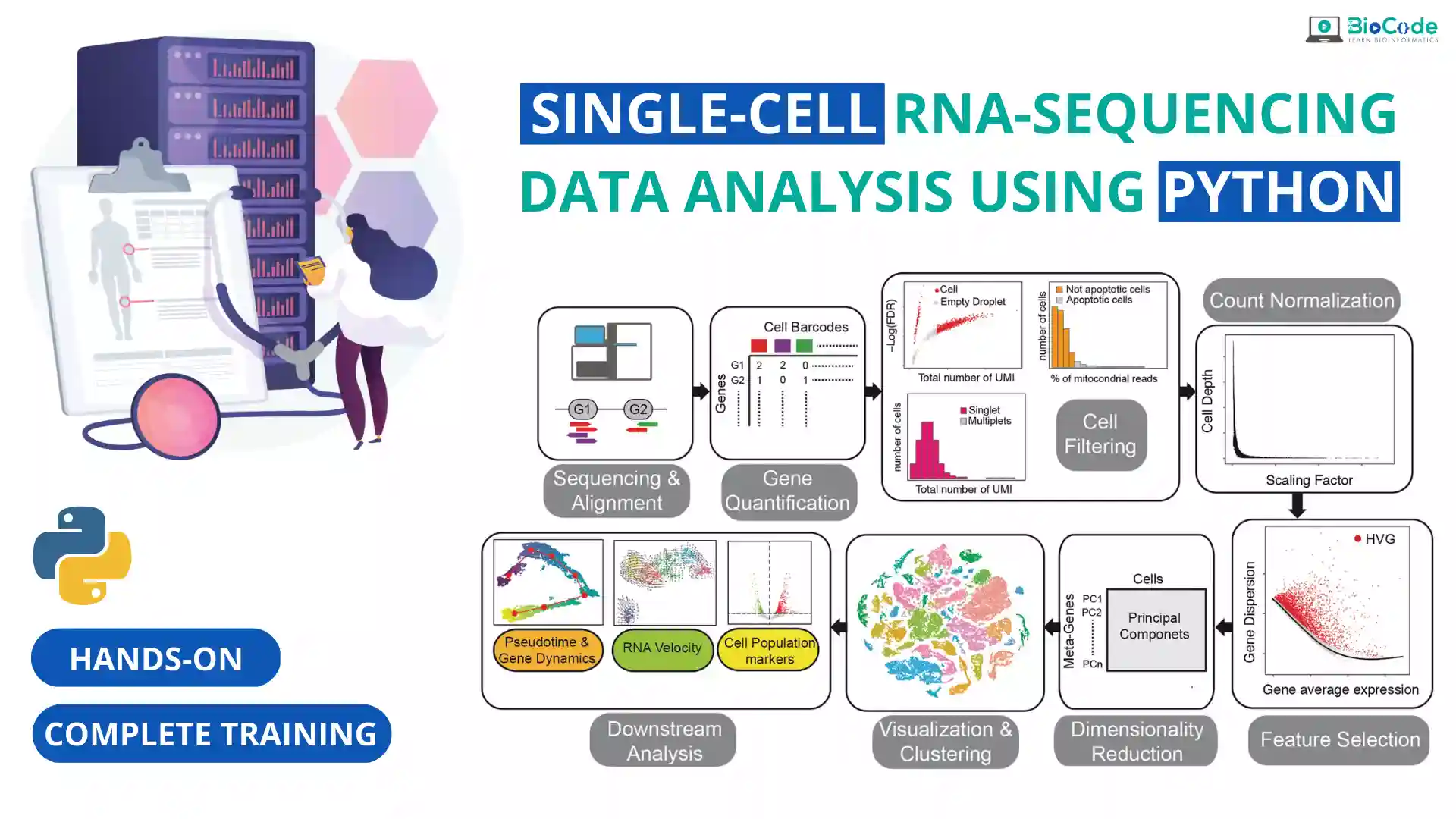
We present a comprehensive single cell RNA-seq data analysis course designed for beginners and advanced researchers in Life Sciences who are interested in exploring the exciting field of single cell genomics. No prior Python programming or Linux knowledge is required. This course is focused on the analysis of scRNA-seq data using command-line tools and Python packages, including Scanpy and other relevant Python packages (see course content section).
Throughout the course, we will explore the differences between scRNA-seq and bulk RNA-seq, and why scRNA-seq is essential for studying cellular and tumor heterogeneity.
This course is ideal for anyone without any prior knowledge of scRNA analysis, as it provides complete guidance and hands-on training from theground up.
We cover all scRNA technologies with an emphasis on 10x Genomics and Smart-seq2. We teach both theory and hands-on experience of each topic in the entire pipeline. We guide students on how to take raw scRNA datasets, install software and Python packages, perform UMI and cell barcode filtering, align against reference genome, and subsequently perform deduplication and quantification (count matrix).
Our course teaches students how to utilize Python programming (ScanPy package) to perform quality control on scRNA data, remove unwanted dead cells that may hinder analysis results, perform normalization, scaling, data imputation, and dimension reduction, and create visualizations through UMAP, tSNE, and PCA plots. We also provide instruction on cell clustering and identifying cell subpopulations in samples, annotating them (cell annotation), and conducting cell lineage and trajectory analysis of the cells.
Finally, we teach students how to analyze differentially expressed genes in either all of the subpopulations or their chosen subpopulations of cells, identifying marker genes that are only expressed in their chosen subpopulations.
By the end of the course, students will have a comprehensive understanding of single cell RNA-seq data analysis using Python packages, and be equipped to tackle their own single-cell sequencing projects.
Course Content
In-depth Introduction to Single Cell RNA-Sequencing, Pipeline and Single-Cell Technologies
-
Introduction to Single-Cell RNA-seq, Its Pipeline and Analysis
23:34 -
Gene Expression and Its Significance
21:42 -
Cellular and Tumor Heterogeneity
09:10 -
Bulk RNA-sequencing vs. Single-Cell RNA-sequencing
21:50 -
Single-Cell RNA-seq Technologies (10x Genomics, Smart-Seq, Drop-seq and more)
23:09 -
Cell Isolation and Cell Lysis Protocols for Single-Cell Genomics
09:53 -
In-depth Introduction to Single-Cell RNA-sequencing Analysis Pipeline
24:45 -
Droplet Technologies: 10x Genomics
14:10 -
Full-Length Transcript Technologies: Smart-Seq2
17:55
Hands-on Single-Cell RNA-seq Data Analysis (From Raw Reads to Cell Subpopulation Identification and DEGs Identification)
Additional Supplementary Lectures
Earn a certificate
Add this certificate to your resume to demonstrate your skills & increase your chances of getting noticed.

Student Ratings & Reviews

