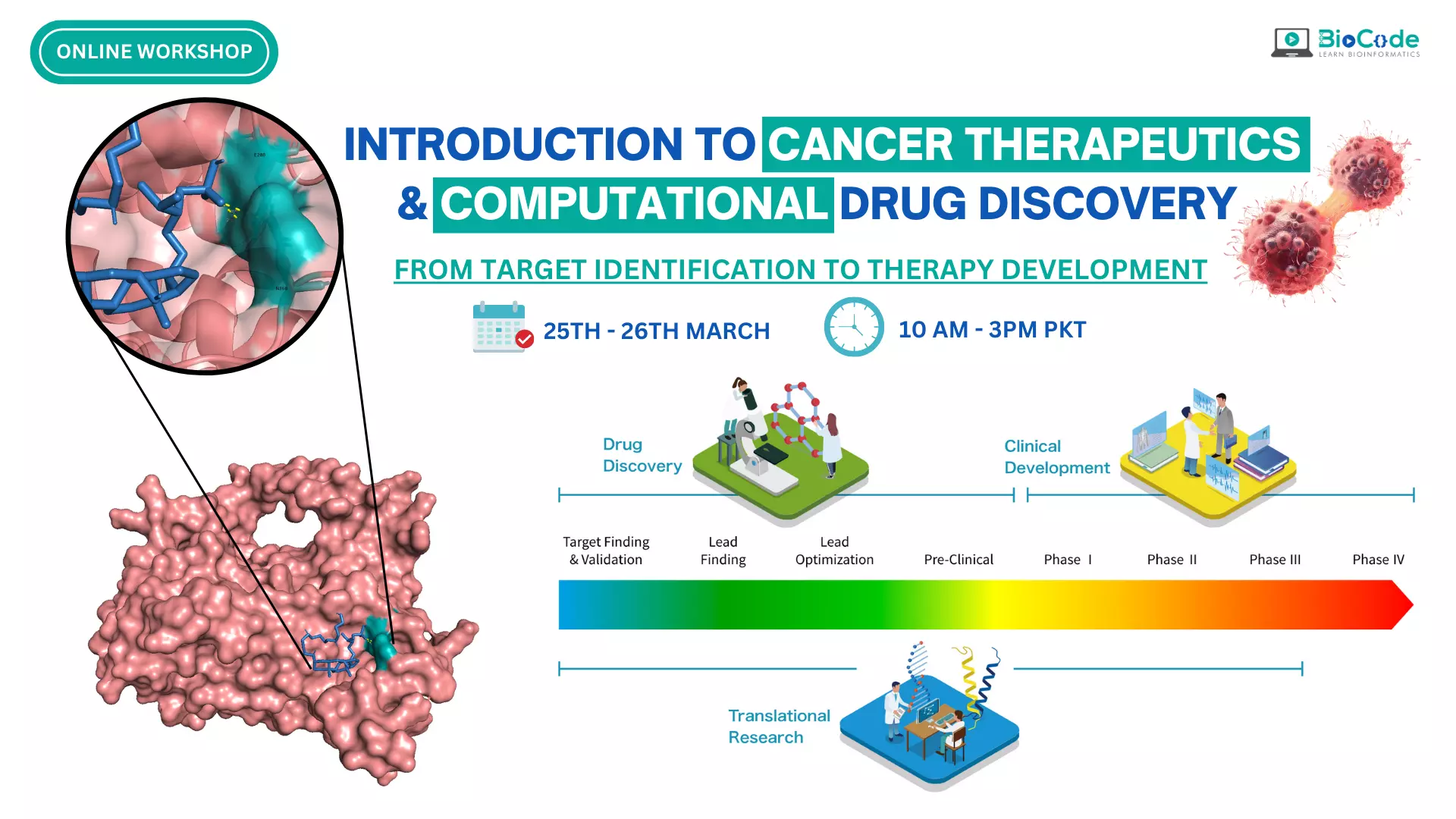
Workshop Cancer Therapeutics & Computational Drug Discovery
About Course
Cancer is a complex disease that affects millions of people worldwide and is one of the leading causes of death globally. The development of effective cancer therapies and drugs is critical in the fight against this disease. Cancer therapeutics are essential in the treatment of cancer. They help to shrink tumors, slow the spread of cancer, and alleviate symptoms such as pain and discomfort. In some cases, cancer treatments can even put cancer into remission, meaning the cancer is no longer detectable in the body.
Computational drug discovery has emerged as a powerful tool in the development of new cancer drugs. Computational tools can help identify potential drug targets, predict the efficacy of new drugs, and accelerate the drug development process. Overall, the importance of cancer therapeutics and drug discovery cannot be overstated. These fields are critical in the fight against cancer, and the development of new and more effective treatments and therapies will continue to improve the lives of cancer patients worldwide.
Therefore, BioCode is offering a hands-on online workshop on introduction to cancer therapeutics and computational drug discovery which is designed to provide an overview of the principles of cancer therapy and how computational drug discovery can be used to identify potential therapeutic targets and develop new drugs. The workshop is aimed at students, researchers, and professionals from all the fields of life sciences for example bioinformatics, medicine, pharmacology, and biotechnology.
The workshop will consist of several sessions, each focusing on a specific topic related to cancer therapeutics and computational drug discovery. The sessions will be conducted by experts in the field and will include lectures, interactive discussions, and hands-on exercises. Throughout the workshop, participants will have the opportunity to interact with the instructor in case of any query or ambiguity. The workshop will also include hands-on exercises to allow participants to apply the concepts learned in the sessions. No prior knowledge or experience is required to attend this workshop.
Upon completion of the workshop, participants will have a better understanding of the principles of cancer therapy and the role of computational drug discovery in developing new treatments. Participants will also have gained practical experience in using computational methods for drug discovery. By studying cancer therapeutics and their effects on cancer cells, researchers can gain insights into the underlying biology of cancer and develop new drugs and treatments that are more effective and targeted. This workshop will provide participants with a broad range of skills and knowledge that can be applied to a variety of research areas in cancer biology and drug discovery.
Session 1: Introduction to Cancer Biology
Description: In this session, participants will learn about the basic principles of cancer biology, including the molecular mechanisms of cancer initiation and progression. The session will cover the hallmarks of cancer, the role of genetic and epigenetic changes in cancer, and the current state of knowledge in cancer biology.
Learning Outcomes: Upon completion of this section, participants will be able to:
- Explain Cancer.
- Discuss Cancer Biology.
- Describe Cancer Initiation and Progression.
- Explain the Role of Genetic and Epigenetic Changes in Cancer.
Session 2: Cancer Metastasis and Resistance
Description: In this session, participants will learn about the mechanisms that underlie cancer metastasis and resistance to treatment. The session will cover the molecular mechanisms of cancer cell invasion, the role of the tumor microenvironment in cancer progression, and the molecular mechanisms of resistance to cancer therapies.
Learning Outcomes: Upon completion of this section, participants will be able to:
- Explain Cancer Metastasis.
- Describe Cancer Cell Invasion.
- Explain the Role of the Tumor Microenvironment in Cancer Progression.
- Describe the Molecular Mechanism of Resistance to Cancer Therapeutics.
Session 3: Introduction to Cancer Therapeutics
Description: In this session, participants will learn about the different types of cancer therapies and their mechanisms of action. The session will cover chemotherapy, radiation therapy, immunotherapy, targeted therapy, hormonal therapy, and the principles and practices of cancer therapy. Participants will be able to identify drug targets for cancer through hands-on guidance.
Learning Outcomes: Upon completion of this section, participants will be able to:
- Explain the Principles and Practices of Cancer Therapy.
- Discuss Chemotherapy.
- Explain Radiation Therapy.
- Describe Immunotherapy.
- Discuss Targeted Therapy.
- Explain Hormonal Therapy.
- Identify Diseases for Novel Research.
- Identify Drug Targets for the Disease.
- Discover Drug Targets for Cancer.
Session 4: Computational Drug Discovery
Description: In this session, participants will learn about computational drug discovery and its importance in developing new cancer treatments. Participants will learn about the principles and practices of computational drug discovery, including molecular modeling and virtual screening. Participants will gain hands-on experience with molecular docking and virtual screening. Participants will also learn about the various types of molecular docking.
Learning Outcomes: Upon completion of this section, participants will be able to:
- Explain Computational Drug Discovery.
- Discuss the Importance of Computational Drug Discovery.
- Describe the Principles and Practices of Computational Drug Discovery.
- Perform End-to-End Molecular Docking.
- Perform End-to-End Virtual Screening Against the Library of Compounds.
- Explain Protein-Ligand Docking.
- Explain Protein-Protein Docking.
- Explain Protein-Peptide Docking.
- Discuss Rigid Body Docking.
- Describe Rigid Receptor – Flexible Ligand Docking.
- Discuss Flexible Receptor – Flexible Ligand Docking.
- Explain Blind Docking.
Session 5: Identification of Drug Targets for Cancer
Description: In this session, participants will learn how to identify potential drug targets for the treatment of cancer using computational tools. Participants will gain practical experience in working with real-world datasets and using computational tools to identify potential therapeutic targets.
Learning Outcomes: Upon completion of this section, participants will be able to:
- Identify Drug Targets for Cancer.
Session 6: 3D Modeling Using Multiple Techniques
Description: In this session, participants will learn about the different techniques used for the 3D structure prediction of proteins and ligands. The session will cover the principles and practices of 3D structure visualization, including molecular graphics, surface representation, and molecular animation. Participants will learn about the tools and methods that are used for 3D structure prediction using ab initio, homology, and threading.
Learning Outcomes: Upon completion of this section, participants will be able to:
- Explain 3D Structure Prediction.
- Perform 3D structure Prediction Using MODELLER.
- Perform 3D structure Prediction Using OmegaFold.
- Perform 3D Structure Prediction Using Ab-initio Method.
- Perform 3D Structure Prediction Using Homology Modelling.
- Perform 3D Structure Prediction Using Threading.
Session 7: Identification of Drug Compounds, Ligand Preparation, Molecular Docking, and Virtual Screening
Description: In this session, participants will learn about the principles and practices of ligand preparation, molecular docking, and virtual screening in drug discovery. In this session, participants will also learn how to retrieve chemical compounds for the drug target to prepare drug libraries for molecular docking. Participants will gain practical experience in using computational tools to identify potential drug compounds and perform virtual screening of potential drug candidates.
Learning Outcomes: Upon completion of this section, participants will be able to:
- Retrieve Drug Compounds from ChEMBL, DrugBank, PubChem, and ZINC.
- Explain Ligand Preparation.
- Perform Ligand Preparation Using AutoDock Vina.
- Perform Molecular Docking Using AutoDock Vina.
- Perform Virtual Screening Using AutoDock Vina.
Session 8: ADMET, Post-Docking Analysis and Visualization of the Docked Complex
Description: In this session, participants will learn that after a candidate compound has been identified through molecular docking or other screening techniques, ADMET properties are evaluated in order to identify it as a potential drug candidate. Participants will learn about the principles and practices of post-docking analysis, including the analysis of binding modes, the calculation of binding energies, and the assessment of drug likeness. In the end, participants will visualize the protein-ligand docked complex in order to see the protein-ligand interactions and generate high-quality publication-ready figures.
Learning Outcomes: Upon completion of this section, participants will be able to:
- Evaluate the ADMET Properties of the Drug Candidate Using SwissADME.
- Perform Post-Docking Analysis.
- Analyze Binding Modes.
- Calculate Binding Energies.
- Assess Drug-Likeness.
- Perform 3D Structure Visualization of the Protein-Ligand Docked Complex on PyMOL and Chimera.
Session 9: Molecular Dynamics and Simulation of Protein-Ligand Complexes
Description: In this session, participants will learn how to perform molecular dynamics and simulation of protein-ligand complexes. The session will cover the basics of molecular dynamics simulations, including force fields, molecular mechanics, and conformational sampling. Participants will learn how to interpret the results that are gained from molecular dynamics simulations.
Learning Outcomes: Upon completion of this section, participants will be able to:
- Explain the Purpose of Molecular Dynamic Simulations.
- Perform Molecular Dynamics Simulation Using Desmond and GROMACS.
- Interpret the RMSD Analysis Results.
- Interpret the RMSF Analysis Results.
- Analyze the Protein-Ligand and Ligand-Protein Contacts.

Course Content
Hands-on Cancer Therapeutics
-
Introduction To Cancer Biology, Cancer Types And Abnormal Cell Cycles
18:17 -
Cancer Initiation: A Cascade Of Abnormal Processes
05:14 -
Cancer Progression Growth, Invasion & Spread
06:44 -
Cancer Metastasis
03:49 -
Cancer Treatments
04:51 -
Identification of Novel Drug Targets
35:46 -
Complete MasterClass on Computational Drug Discovery and Molecular Docking
50:14 -
Retrieval of Target Protein and 3D Structure Predeiction Methods
25:41 -
Loop Modeling – Fixing the Unmodelled Regions in Target Proteins
19:22
Excerice
Earn a certificate
Add this certificate to your resume to demonstrate your skills & increase your chances of getting noticed.

My suggestion for next topic is virtual screening of anticancer drugs using molecular docking.the second part should be dealt with the target protein in cancer. Prepare the protein by discovery studio and screening for anticancer drugs using pyrx etc and molecular dynamics simulation.
That would be a full class.hope biocode built upon second part