Cancer Genomics: NGS (Whole Genome Sequencing) Variant Calling Using Linux
About Course
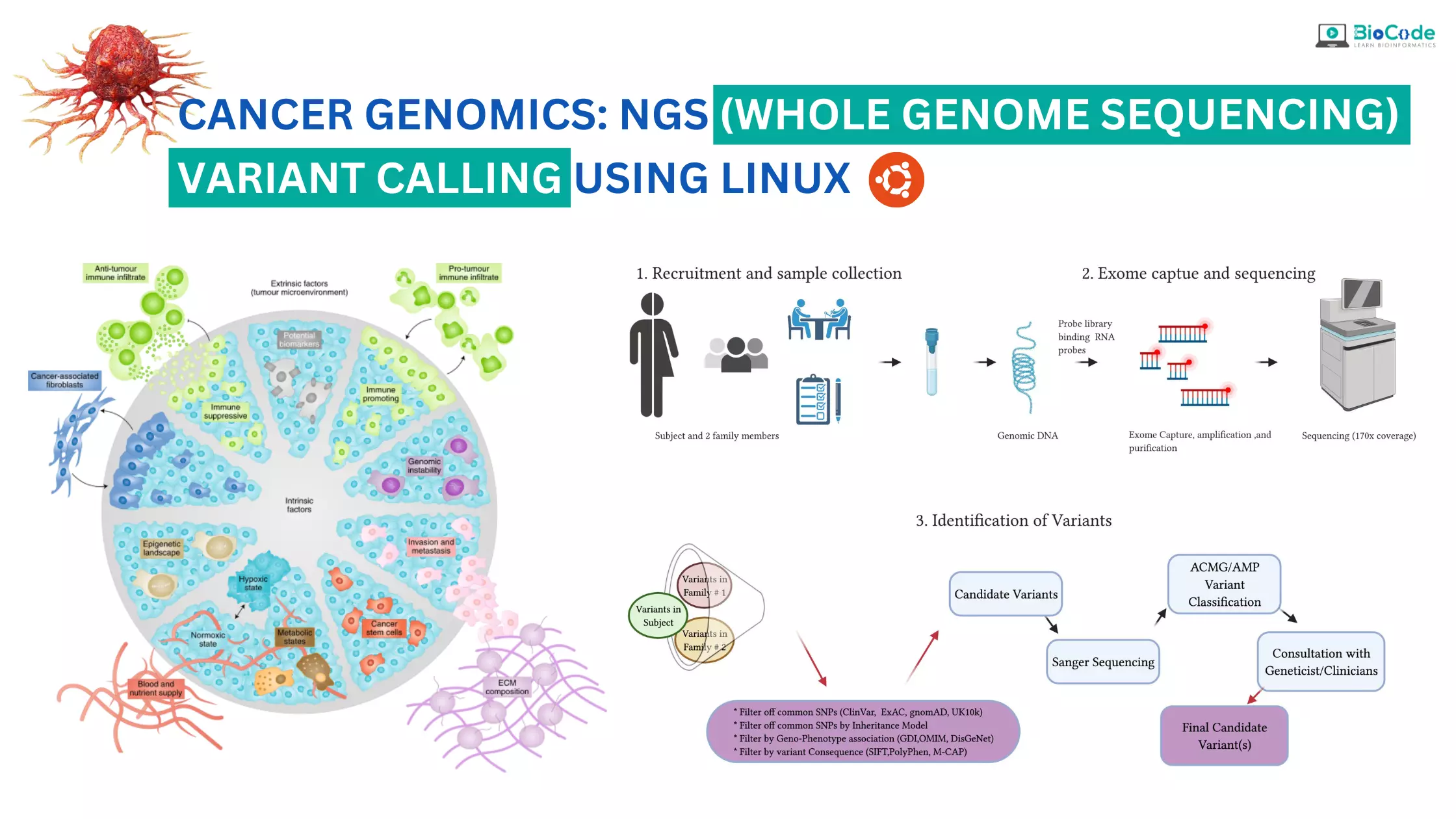
In the field of cancer genomics, the analysis of NGS/whole genome data has become an integral part of cancer research. This course provides a comprehensive overview of the process of analyzing NGS data to identify genetic variants in cancer patients, including substitutions (SNPs) and insertions/deletions (indels).
The course begins by covering the basics of NGS, including the technologies and sequencing platforms used in NGS experiments. It then moves on to teach the student how to obtain raw whole genome datasets for any organism of any disease, and how to perform quality control on these datasets.
The student will learn how to preprocess the data, including trimming and mapping the reads against a reference genome. They will also learn about post-alignment quality control and the process of recalibrating reads to ensure accuracy.
The main focus of the course is on variant calling, where the student will learn how to identify variants in the disease/patient samples using various tools and algorithms. This process involves identifying and classifying genetic variants, including SNPs and indels, and annotating them to determine their functional significance.
The student will also learn how to filter and annotate the resulting variant call format (VCF) files to obtain meaningful insights into the genetic basis of disease. These annotations may include functional annotations such as gene
annotations, pathway annotations, and annotations related to the frequency of the variant in the general population.
Overall, this course provides a comprehensive introduction to NGS data analysis for cancer genomics, including the process of obtaining, processing, and analyzing whole genome data to identify genetic variants that may be associated with cancer.
Course Content
In-depth Introduction to NGS and Whole-Genome Sequencing for Cancer Genomics
-
In-depth Introduction to NGS and Variant Calling
20:12 -
Fundamentals of Whole Genome Sequencing: A Comprehensive Approach to Sequence a Genome
00:00 -
Cancer Genomics: Role of WGS in Cancer Research
00:00 -
Haploid vs. Diploid Organisms for Variant Calling
07:26 -
Ploidy and Its Significance in Disease Research
14:58 -
A Bioinformatics Approach Towards Germline vs. Somatic Mutations
14:30 -
Single Nucleotide Polymorphisms: An Introduction to SNPs and Their Types
31:22 -
Structural Variations: Large Scale Variations in Genomes
08:55 -
Copy Number Variations: Role of CNVs in Diseases
10:29
Hands-on Whole-Genome Variant Calling
Additional Downstream Analysis
Additional Lectures
Earn a certificate
Add this certificate to your resume to demonstrate your skills & increase your chances of getting noticed.

Student Ratings & Reviews

